MicroRNAs as Potential Biomarkers in Atherosclerosis
Abstract
:1. Introduction
2. Biological Role of miRNAs
MiRNA Biogenesis
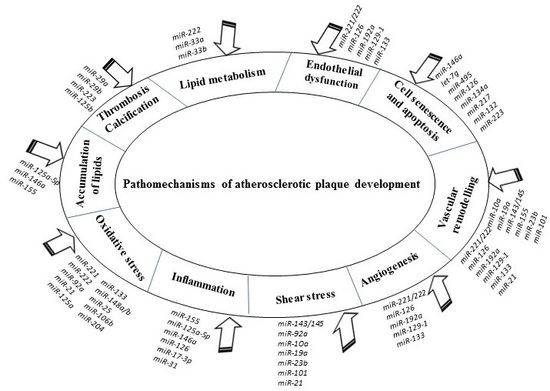
3. Roles of Various miRNAs in Developmental Pathomechanisms of Atherosclerosis
3.1. miRNAs in the Regulation of Endothelial Cell Function
3.2. miRNAs in the Regulation of Endothelial Cell Senescence
3.3. miRNAs in Regulation of Endothelial Cell Apoptosis
3.4. miRNAs in the Regulation of Lipid Retention and Local Inflammation
3.5. miRNAs in the Regulation of VSMC Function
3.6. MiRNAs in the Regulation of Calcification in VSMCs
3.7. miRNAs in the Regulation of Cholesterol Metabolism
4. MiRNAs as Diagnostic Biomarkers in Atherosclerosis and CVDs
5. RNA-Based Therapeutic Approaches for the Treatment of Atherosclerosis and CVDs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Orekhov, A.N.; Ivanova, E.A. Introduction of the special issue “Atherosclerosis and Related Diseases”. Vessel. Plus 2017, 1, 163–165. [Google Scholar] [CrossRef]
- Alipov, V.I.; Sukhorukov, V.N.; Karagodin, V.P.; Grechko, A.V.; Orekhov, A.N. Chemical composition of circulating native and desialylated low density lipoprotein: What is the difference? Vessel. Plus 2017. [Google Scholar] [CrossRef]
- Hansson, G.K.; Robertson, A.K.; Söderberg-Nauclér, C. Inflammation and atherosclerosis. Annu. Rev. Pathol.: Mech. Dis. 2006, 1, 297–329. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Maseri, A. Inflammation and Atherosclerosis. Circulation 2002, 105, 1135–1143. [Google Scholar] [CrossRef]
- Packard, R.R.S.; Lichtman, A.H.; Libby, P. Innate and adaptive immunity in atherosclerosis. Semin. Immunopathol. 2009, 31, 5–22. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Hsu, P.-P.; Chen, B.P.; Yuan, S.; Usami, S.; Shyy, J.Y.-J.; Li, Y.-S.; Chien, S. Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc. Natl. Acad. Sci. USA 2000, 97, 9385–9389. [Google Scholar] [CrossRef] [Green Version]
- Cerne, A.; Kranjec, I. Atherosclerotic burden in coronary and peripheral arteries in patients with first clinical manifestation of coronary artery disease. Hear. Vessel. 2002, 16, 217–226. [Google Scholar] [CrossRef]
- Eikendal, A.L.M.; Groenewegen, K.A.; Bots, M.L.; Peters, S.A.E.; Uiterwaal, C.S.P.M.; den Ruijter, H.M. Relation Between Adolescent Cardiovascular Risk Factors and Carotid Intima-Media Echogenicity in Healthy Young Adults: The Atherosclerosis Risk in Young Adults (ARYA) Study. J. Am. Heart Assoc. 2019, 5, e002941. [Google Scholar] [CrossRef]
- Insull, W. The Pathology of Atherosclerosis: Plaque Development and Plaque Responses to Medical Treatment. Am. J. Med. 2009, 122, S3–S14. [Google Scholar] [CrossRef]
- Kumar, S.; Verma, A.K.; Kumar, N.; Verma, R.K. Prevalence of Coronary Atherosclerosis in Different Age Groups: A Postmor- tem Study. Biomed. Res. 2013, 24, 5. [Google Scholar]
- Mcmahan, C.A.; Fayad, Z.A.; Zieske, A.W.; Gidding, S.S.; Malcom, G.T.; Tracy, R.E.; Strong, J.P.; McGill, H.C. Risk Scores Predict Atherosclerotic Lesions in Young People. Arch. Intern. Med. 2005, 165, 883–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spring, B.; Moller, A.C.; Colangelo, L.A.; Siddique, J.; Roehrig, M.; Daviglus, M.L.; Polak, J.F.; Reis, J.P.; Sidney, S.; Liu, K. Healthy Lifestyle Change and Subclinical Atherosclerosis in Young Adults. Circulation 2014, 130, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuzcu, E.M.; Kapadia, S.R.; Tutar, E.; Ziada, K.M.; Hobbs, R.E.; McCarthy, P.M.; Young, J.B.; Nissen, S.E. High Prevalence of Coronary Atherosclerosis in Asymptomatic Teenagers and Young Adults. Circulation 2001, 103, 2705–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzou, W.; Douglas, P.; Srinivasan, S. Increased Subclinical Atherosclerosis in Young Adults with Metabolic Syndrome. The Bogalusa Heart Study. ACC Curr. J. Rev. 2005, 14, 18. [Google Scholar] [CrossRef]
- Wang, J.; Niu, D.; Meng, Y.; Han, A.; Li, K.; Zhang, C. Plasma oxidized lipoprotein(a) and its immune complexes are present in newborns and children. Clin. Chim. Acta 2009, 407, 1–5. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and non-conserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Ying, S.Y.; Lin, S.L. Current perspectives in intronic micro RNAs (miRNAs). J. Biomed. Sci. 2006, 13, 5–15. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- De Rosa, S.; Curcio, A.; Indolfi, C. Emerging Role of MicroRNAs in Cardiovascular Diseases. Circ. J. 2014, 78, 567–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Mezzetti, A.; Cipollone, F. MicroRNAs and atherosclerosis: New actors for an old movie. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 868–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacomino, G.; Siani, A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Shirafkan, N.; Mansoori, B.; Mohammadi, A.; Shomali, N.; Ghasbi, M.; Baradaran, B. MicroRNAs as novel biomarkers for colorectal cancer: New outlooks. Biomed. Pharmacother. 2018, 97, 1319–1330. [Google Scholar] [CrossRef]
- Tang, X.; Tang, G.; Özcan, S. Role of microRNAs in diabetes. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2008, 1779, 697–701. [Google Scholar] [CrossRef] [Green Version]
- Daugaard, I.; Hansen, T.B. Biogenesis and Function of Ago-Associated RNAs. Trends Genet. 2017, 33, 208–219. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004, 14, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs Exhibit Strand Bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.S. How to slice: Snapshots of Argonaute in action. Silence 2010, 1, 3. [Google Scholar] [CrossRef]
- Lee, Y.; Hur, I.; Park, S.-Y.; Kim, Y.-K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Chang, K.-W.; Kao, S.-Y.; Wu, Y.-H.; Tsai, M.-M.; Tu, H.-F.; Liu, C.-J.; Lui, M.-T.; Lin, S.-C. Passenger strand miRNA miR-31∗ regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013, 49, 27–33. [Google Scholar] [CrossRef]
- Yang, J.-S.; Phillips, M.D.; Betel, D.; Mu, P.; Ventura, A.; Siepel, A.C.; Chen, K.C.; Lai, E.C. Widespread regulatory activity of vertebrate microRNA* species. RNA 2011, 17, 312–326. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Siomi, H. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol. Genet. Genom. 2010, 284, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genome Res. 2006, 20, 2202–2207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez, Y.; Fernández-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for Endothelial MicroRNA Expression and Angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Houbaviy, H.B.; Murray, M.F.; A Sharp, P. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Mitomo, S.; Maesawa, C.; Ogasawara, S.; Iwaya, T.; Shibazaki, M.; Yashima-Abo, A.; Kotani, K.; Oikawa, H.; Sakurai, E.; Izutsu, N.; et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008, 99, 280–286. [Google Scholar] [CrossRef]
- Obernosterer, G.; Leuschner, P.J.; Alenius, M.; Martinez, J. Post-transcriptional regulation of microRNA expression. RNA 2006, 12, 1161–1167. [Google Scholar] [CrossRef] [Green Version]
- Chamorro-Jorganes, A.; Araldi, E.; Suárez, Y. MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol. Res. 2013, 75, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Belkin, N.; Feinberg, M.W. Endothelial microRNAs and atherosclerosis. Curr. Atheroscler. Rep. 2013, 15, 372. [Google Scholar] [CrossRef]
- Zhang, X.; Mao, H.; Chen, J.-Y.; Wen, S.; Li, D.; Ye, M.; Lv, Z. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem. Biophys. Res. Commun. 2013, 431, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Donovan, P.; Khosrotehrani, K. Concise Review: Functional Definition of Endothelial Progenitor Cells: A Molecular Perspective. STEM CELLS Transl. Med. 2016, 5, 1302–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampetaki, A.; Kirton, J.P.; Xu, Q. Vascular repair by endothelial progenitor cells. Cardiovasc. Res. 2008, 78, 413–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuehbacher, A.; Urbich, C.; Dimmeler, S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol. Sci. 2008, 29, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Suarez, Y.; Fernaández-Hernando, C.; Pober, J.S.; Sessa, W.C.; Suárez, Y.; Fernández-Hernando, C. Dicer Dependent MicroRNAs Regulate Gene Expression and Functions in Human Endothelial Cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [Green Version]
- Fish, J.E.; Marsden, P.A. Endothelial nitric oxide synthase: Insight into cell-specific gene regulation in the vascular endothelium. Cell. Mol. Life Sci. 2006, 63, 144–162. [Google Scholar] [CrossRef]
- Incalza, M.A.; D’Oria, R.; Natalicchio, A.; Perrini, S.; Laviola, L.; Giorgino, F. Oxidative stress and reactive oxygen species in endothelial dysfunction associated with cardiovascular and metabolic diseases. Vasc. Pharmacol. 2018, 100, 1–19. [Google Scholar] [CrossRef]
- Rudic, R.D.; Shesely, E.G.; Maeda, N.; Smithies, O.; Segal, S.S.; Sessa, W.C. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J. Clin. Investig. 1998, 101, 731–736. [Google Scholar] [CrossRef]
- Ziche, M.; Morbidelli, L.; Masini, E.; Amerini, S.; Granger, H.J.; A Maggi, C.; Geppetti, P.; Ledda, F. Nitric oxide mediates angiogenesis in vivo and endothelial cell growth and migration in vitro promoted by substance P. J. Clin. Investig. 1994, 94, 2036–2044. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soufi-Zomorrod, M.; Hajifathali, A.; Kouhkan, F.; Mehdizadeh, M.; Rad, S.M.A.H.; Soleimani, M. MicroRNAs modulating angiogenesis: miR-129-1 and miR-133 act as angio-miR in HUVECs. Tumor Biol. 2016, 37, 9527–9534. [Google Scholar] [CrossRef] [PubMed]
- Nazari-Jahantigh, M.; Wei, Y.; Schober, A. The role of microRNAs in arterial remodelling. Thromb. Haemost. 2012, 107, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Neth, P.; Nazari-Jahantigh, M.; Schober, A.; Weber, C. MicroRNAs in flow-dependent vascular remodelling. Cardiovasc. Res. 2013, 99, 294–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boon, R.A.; Hergenreider, E.; Dimmeler, S. Atheroprotective mechanisms of shear stress-regulated microRNAs. Thromb. Haemost. 2012, 108, 616–620. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, K.-C.; Wu, W.; Subramaniam, S.; Shyy, J.Y.-J.; Chiu, J.-J.; Li, J.Y.-S.; Chien, S. MicroRNA-21 targets peroxisome proliferators-activated receptor-α in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc. Natl. Acad. Sci. USA 2011, 108, 10355–10360. [Google Scholar] [CrossRef]
- Weber, M.; Baker, M.B.; Moore, J.P.; Searles, C.D. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem. Biophys. Res. Commun. 2010, 393, 643–648. [Google Scholar] [CrossRef] [Green Version]
- Varga, Z.V.; Kupai, K.; Szűcs, G.; Gáspár, R.; Pálóczi, J.; Faragó, N.; Zvara, Á.; Puskás, L.G.; Rázga, Z.; Tiszlavicz, L.; et al. MicroRNA-25-dependent upregulation of NADPH oxidase 4 (NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress and subsequent dysfunction in the heart. J. Mol. Cell. Cardiol. 2013, 62, 111–121. [Google Scholar] [CrossRef]
- Yang, J.; Brown, M.E.; Zhang, H.; Martínez, M.; Zhao, Z.; Bhutani, S.; Yin, S.; Trac, D.; Xi, J.J.; Davis, M.E. High-throughput screening identifies microRNAs that target Nox2 and improve function after acute myocardial infarction. Am. J. Physiol. Circ. Physiol. 2017, 312, H1002–H1012. [Google Scholar] [CrossRef] [Green Version]
- Fleissner, F.; Jazbutyte, V.; Fiedler, J.; Galuppo, P.; Mayr, M.; Ertl, G.; Bauersachs, J.; Thum, T. The endogenous NO synthase inhibitor asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA dependent mechanism. Cardiovasc. Res. 2010, 87, S45–S88. [Google Scholar]
- Zhang, X.; Ng, W.-L.; Wang, P.; Tian, L.; Werner, E.; Wang, H.; Doetsch, P.; Wang, Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012, 72, 4707–4713. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, H.; Wu, J.; Zhao, Y. miR-125a Suppresses TrxR1 Expression and Is Involved in H2O2-Induced Oxidative Stress in Endothelial Cells. J. Immunol. Res. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hu, Y.; Hou, L.; Ju, J.; Li, X.; Du, N.; Guan, X.; Liu, Z.; Zhang, T.; Qin, W.; et al. β-Blocker carvedilol protects cardiomyocytes against oxidative stress-induced apoptosis by upregulating miR-133 expression. J. Mol. Cell. Cardiol. 2014, 75, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Holliday-Ankeny, C.J.; Ankeny, R.F.; Ferdous, Z.; Nerem, R.M.; Jo, H. Shear-and side-dependent microRNAs and messenger RNAs in aortic valvular endothelium. In 5th Biennial Conference on Heart Valve Biology and Tissue Engineering; Hamad bin Khalifa University Press: Doha, Qatar, 2012; p. 56. [Google Scholar] [CrossRef]
- Costantino, S.; Paneni, F.; Cosentino, F. Ageing, metabolism and cardiovascular disease. J. Physiol. 2016, 594, 2061–2073. [Google Scholar] [CrossRef]
- Minamino, T.; Komuro, I. Vascular Cell Senescence. Circ. Res. 2007, 100, 15–26. [Google Scholar] [CrossRef]
- Ito, T.; Yagi, S.; Yamakuchi, M. MicroRNA-34a regulation of endothelial senescence. Biochem. Biophys. Res. Commun. 2010, 398, 735–740. [Google Scholar] [CrossRef]
- Menghini, R.; Casagrande, V.; Cardellini, M.; Martelli, E.; Terrinoni, A.; Amati, F.; Vasa-Nicotera, M.; Ippoliti, A.; Novelli, G.; Melino, G.; et al. MicroRNA 217 Modulates Endothelial Cell Senescence via Silent Information Regulator 1. Circulation 2009, 120, 1524–1532. [Google Scholar] [CrossRef] [Green Version]
- Vasa-Nicotera, M.; Chen, H.; Tucci, P.; Yang, A.L.; Saintigny, G.; Menghini, R.; Mahé, C.; Agostini, M.; Knight, R.A.; Melino, G.; et al. miR-146a is modulated in human endothelial cell with aging. Atherosclerosis 2011, 217, 326–330. [Google Scholar] [CrossRef]
- Liao, Y.-C.; Wang, Y.-S.; Guo, Y.-C.; Lin, W.-L.; Chang, M.-H.; Juo, S.-H.H. Let-7g Improves Multiple Endothelial Functions Through Targeting Transforming Growth Factor-Beta and SIRT-1 Signaling. J. Am. Coll. Cardiol. 2014, 63, 1685–1694. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Sun, Y.; Chen, Y.; Sun, Y.; Li, R.; Liu, C.; Zhang, C.; Wang, R.; Zhang, Y. Endothelial cell apoptosis is responsible for the formation of coronary thrombotic atherosclerotic plaques. Tohoku J. Exp. Med. 2009, 218, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, J.; Wang, B.; Yang, J.; Gong, Z.; Zhao, X.; Zhang, C.; Du, K. MiR-126 inhibits vascular endothelial cell apoptosis through targeting PI3K/Akt signaling. Ann. Hematol. 2016, 95, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, X.-L.; Yan, C.-H.; Li, Y.; Tian, X.-X.; Zhu, N.; Rong, J.-J.; Peng, C.-F.; Han, Y.-L. MicroRNA-495 regulates the proliferation and apoptosis of human umbilical vein endothelial cells by targeting chemokine CCL2. Thromb. Res. 2015, 135, 146–154. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, D.; Wang, Q.; Shen, D.; Wang, Y.; Chen, B.; Zhang, J.; Gai, L. MiR-132 Inhibits Expression of SIRT1 and Induces Pro-inflammatory Processes of Vascular Endothelial Inflammation through Blockade of the SREBP-1c Metabolic Pathway. Cardiovasc. Drugs Ther. 2014, 28, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liang, H.; Liu, H.; Li, D.; Chen, X.; Li, L.; Zhang, C.-Y.; Zen, K. Platelet-Secreted MicroRNA-223 Promotes Endothelial Cell Apoptosis Induced by Advanced Glycation End Products via Targeting the Insulin-like Growth Factor 1 Receptor. J. Immunol. 2014, 192, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tian, F.; Wang, J.; Jing, J.; Zhou, S.-S.; Chen, Y.-D. Endothelial Cell Autophagy in Atherosclerosis is Regulated by miR-30-Mediated Translational Control of ATG6. Cell. Physiol. Biochem. 2015, 37, 1369–1378. [Google Scholar] [CrossRef]
- Yang, K.; He, Y.S.; Wang, X.Q.; Lu, L.; Chen, Q.J.; Liu, J.; Sun, Z.; Shen, W.F. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 2011, 585, 854–860. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Huang, Z.; Wang, L.; Wu, F.; Meng, S. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc. Res. 2009, 83, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Olkkonen, V.M. Oxysterol binding protein and its homologues: New regulatory factors involved in lipid metabolism. Curr. Opin. Lipidol. 2004, 15, 321–327. [Google Scholar] [CrossRef]
- Li, X.; Kong, D.; Chen, H.; Liu, S.; Hu, H.; Wu, T.; Wang, J.; Chen, W.; Ning, Y.; Li, Y.; et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci. Rep. 2016, 6, 21789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, H.; Wang, F.; Zhang, W.; Wang, D.; Li, X.; Bartlam, M.; Yao, X.; Rao, Z. Structure-Functional Analyses of CRHSP-24 Plasticity and Dynamics in Oxidative Stress Response. J. Biol. Chem. 2011, 286, 9623–9635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Chen, T.; Yang, L.; Li, Z.; Wong, M.M.; Zheng, X.; Pan, X.; Zhang, L.; Yan, H. Regulation of MicroRNA-155 in Atherosclerotic Inflammatory Responses by Targeting MAP3K10. PLoS ONE 2012, 7, e46551. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.-Z.; Fan, H.-M. Identification of miRNAs as atherosclerosis biomarkers and functional role of miR-126 in atherosclerosis progression through MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2725–2733. [Google Scholar] [PubMed]
- Yang, Y.; Yang, L.; Liang, X.; Zhu, G. MicroRNA-155 Promotes Atherosclerosis Inflammation via Targeting SOCS1. Cell. Physiol. Biochem. 2015, 36, 1371–1381. [Google Scholar] [CrossRef]
- Yuan, Z.; Guan, Y.; Wang, L.; Wei, W.; Kane, A.B.; Chin, Y.E. Central Role of the Threonine Residue within the p+1 Loop of Receptor Tyrosine Kinase in STAT3 Constitutive Phosphorylation in Metastatic Cancer Cells. Mol. Cell. Biol. 2004, 24, 9390–9400. [Google Scholar] [CrossRef] [Green Version]
- Suárez, Y.; Wang, C.; Manes, T.D.; Pober, J.S. Cutting Edge: TNF-Induced MicroRNAs Regulate TNF-Induced Expression of E-Selectin and Intercellular Adhesion Molecule-1 on Human Endothelial Cells: Feedback Control of Inflammation. J. Immunol. 2010, 184, 21–25. [Google Scholar] [CrossRef]
- Muto, A.; Fitzgerald, T.N.; Pimiento, J.M.; Maloney, S.P.; Teso, D.; Paszkowiak, J.J.; Westvik, T.S.; Kudo, F.A.; Nishibe, T.; Dardik, A. Smooth muscle cell signal transduction: Implications of vascular biology for vascular surgeons. J. Vasc. Surg. 2007, 45, A15–A24. [Google Scholar] [CrossRef] [Green Version]
- Rivard, A.; Andrés, V. Vascular smooth muscle cell proliferation in the pathogenesis of atherosclerotic cardiovascular diseases. Histol. Histopathol. 2000, 15, 557–571. [Google Scholar]
- Ahmed, S.; Warren, D.T. Vascular smooth muscle cell contractile function and mechanotransduction. Vessel. Plus 2018, 2, 36. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Li, G. Role of Specific MicroRNAs in Regulation of Vascular Smooth Muscle Cell Differentiation and the Response to Injury. J. Cardiovasc. Transl. Res. 2010, 3, 246–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, R.; Cheng, Y.; Yue, J.; Yang, J.; Liu, X.; Chen, H.; Dean, D.B.; Zhang, C. MicroRNA Expression Signature and Antisense-Mediated Depletion Reveal an Essential Role of MicroRNA in Vascular Neointimal Lesion Formation. Circ. Res. 2007, 100, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, X.; Yang, J.; Lin, Y.; Xu, D.-Z.; Lu, Q.; Deitch, E.A.; Huo, Y.; Delphin, E.S.; Zhang, C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 2009, 105, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Albinsson, S.; Suarez, Y.; Skoura, A.; Offermanns, S.; Miano, J.M.; Sessa, W.C. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arter. Thromb. Vasc. Biol. 2010, 30, 1118–1126. [Google Scholar] [CrossRef]
- Zhang, C. MicroRNA and vascular smooth muscle cell phenotype: New therapy for atherosclerosis? Genome Med. 2009, 1, 85. [Google Scholar] [CrossRef]
- Zhang, C. MicroRNA-145 in vascular smooth muscle cell biology: A new therapeutic target for vascular disease. Cell Cycle 2009, 8, 3469–3473. [Google Scholar] [CrossRef]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Krüger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Investig. 2009, 119, 2634–2647. [Google Scholar] [CrossRef] [Green Version]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.-H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Xin, M.; Small, E.M.; Sutherland, L.B.; Qi, X.; McAnally, J.; Plato, C.F.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genome Res. 2009, 23, 2166–2178. [Google Scholar] [CrossRef] [Green Version]
- Elia, L.; Quintavalle, M.; Zhang, J.; Contu, R.; Cossu, L.; Latronico, M.V.G.; Peterson, K.L.; Indolfi, C.; Catalucci, D.; Chen, J.; et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009, 16, 1590–1598. [Google Scholar] [CrossRef]
- Torella, D.; Iaconetti, C.; Catalucci, D.; Ellison, G.M.; Leone, A.; Waring, C.D.; Bochicchio, A.; Vicinanza, C.; Aquila, I.; Curcio, A.; et al. MicroRNA-133 Controls Vascular Smooth Muscle Cell Phenotypic Switch In Vitro and Vascular Remodeling In Vivo. Circ. Res. 2011, 109, 880–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Bezprozvannaya, S.; Williams, A.H.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genome Res. 2008, 22, 3242–3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Hu, X.; Zhang, Q.; Wang, J.; Li, J.; Liu, B.; Shao, Y.; Li, X.; Zhang, J.; Xin, S. Upregulation of let-7a inhibits vascular smooth muscle cell proliferation in vitro and in vein graft intimal hyperplasia in rats. J. Surg. Res. 2014, 192, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.-L.; Wang, J.-F.; Wang, G.-K.; You, X.-H.; Zhao, X.-X.; Jing, Q.; Qin, Y.-W. Vascular smooth muscle cell proliferation is influenced by let-7d microRNA and its interaction with KRAS. Circ. J. 2011, 75, 703–709. [Google Scholar] [CrossRef]
- Chen, K.-C.; Hsieh, I.-C.; Hsi, E.; Wang, Y.-S.; Dai, C.-Y.; Chou, W.-W.; Juo, S.-H.H. Negative feedback regulation between microRNA let-7g and the oxLDL receptor LOX-1. J. Cell Sci. 2011, 124, 4115–4124. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cheng, Y.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ. Res. 2009, 104, 476–487. [Google Scholar] [CrossRef]
- Du, Y.; Gao, C.; Liu, Z.; Wang, L.; Liu, B.; He, F.; Zhang, T.; Wang, Y.; Xu, M.; Luo, G.-Z.; et al. Upregulation of a Disintegrin and Metalloproteinase with Thrombospondin Motifs-7 by miR-29 Repression Mediates Vascular Smooth Muscle Calcification. Arter. Thromb. Vasc. Biol. 2012, 32, 2580–2588. [Google Scholar] [CrossRef]
- Wen, P.; Cao, H.; Fang, L.; Ye, H.; Zhou, Y.; Jiang, L.; Su, W.; Xu, H.; He, W.; Dai, C.; et al. miR-125b/Ets1 axis regulates transdifferentiation and calcification of vascular smooth muscle cells in a high-phosphate environment. Exp. Cell Res. 2014, 322, 302–312. [Google Scholar] [CrossRef]
- Goettsch, C.; Rauner, M.; Pacyna, N.; Hempel, U.; Bornstein, S.R.; Hofbauer, L.C. miR-125b Regulates Calcification of Vascular Smooth Muscle Cells. Am. J. Pathol. 2011, 179, 1594–1600. [Google Scholar] [CrossRef]
- Rayner, K.J.; Fernández-Hernando, C.; Moore, K.J. MicroRNAs regulating lipid metabolism in atherogenesis. Thromb. Haemost. 2012, 107, 642–647. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suárez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Clerbaux, L.-A.; Haumont, O.; Lanthier, N.; Das, A.K.; Burant, C.F.; Leclercq, I.A.; MacDougald, O.A.; Bommer, G.T. Expression of miR-33 from an SREBP2 Intron Inhibits Cholesterol Export and Fatty Acid Oxidation*. J. Biol. Chem. 2010, 285, 33652–33661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquart, T.J.; Allen, R.M.; Ory, D.S.; Baldán, Á. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 12228–12232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tall, A.R.; Yvan-Charvet, L.; Terasaka, N.; Pagler, T.; Wang, N. HDL, ABC Transporters, and Cholesterol Efflux: Implications for the Treatment of Atherosclerosis. Cell Metab. 2008, 7, 365–375. [Google Scholar] [CrossRef] [Green Version]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Elmen, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.W.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef]
- Urbich, C.; Kaluza, D.; Frömel, T.; Knau, A.; Bennewitz, K.; Boon, R.A.; Bonauer, A.; Doebele, C.; Boeckel, J.-N.; Hergenreider, E.; et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 2012, 119, 1607–1616. [Google Scholar] [CrossRef] [Green Version]
- Poissonnier, L.; Villain, G.; Soncin, F.; Mattot, V. miR126-5p repression of ALCAM and SetD5 in endothelial cells regulates leucocyte adhesion and transmigration. Cardiovasc. Res. 2014, 102, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Schober, A.; Nazari-Jahantigh, M.; Wei, Y.; Bidzhekov, K.; Gremse, F.; Grommes, J.; A Megens, R.T.; Heyll, K.; Noels, H.; Hristov, M.; et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat. Med. 2014, 20, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Köppel, T.; Jahantigh, M.N.; Lutgens, E.; et al. Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12-Dependent Vascular Protection. Sci. Signal. 2009, 2, ra81. [Google Scholar] [CrossRef] [PubMed]
- Meiler, S.; Baumer, Y.; Toulmin, E.; Seng, K.; Boisvert, W.A. MicroRNA 302a Is a Novel Modulator of Cholesterol Homeostasis and Atherosclerosis. Arter. Thromb. Vasc. Biol. 2015, 35, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, C.M.; Davalos, A.; Goedeke, L.; Salerno, A.G.; Warrier, N.; Cirera-Salinas, D.; Suárez, Y.; Fernández-Hernando, C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arter. Thromb. Vasc. Biol. 2011, 31, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Nazari-Jahantigh, M.; Chan, L.; Zhu, M.; Heyll, K.; Corbalán-Campos, J.; Hartmann, P.; Thiemann, A.; Weber, C.; Schober, A. The microRNA-342-5p Fosters Inflammatory Macrophage Activation Through an Akt1- and microRNA-155-Dependent Pathway During Atherosclerosis. Circulation 2013, 127, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1α and affects lipid metabolism[S]. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shi, C.; Manduchi, E.; Civelek, M.; Davies, P.F. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 13450–13455. [Google Scholar] [CrossRef] [Green Version]
- Ni, C.-W.; Qiu, H.; Jo, H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am. J. Physiol. Circ. Physiol. 2011, 300, H1762–H1769. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhu, N.; Yi, B.; Wang, N.; Chen, M.; You, X.; Zhao, X.; Solomides, C.C.; Qin, Y.; Sun, J. MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation. Circ. Res. 2013, 113, 1117–1127. [Google Scholar] [CrossRef]
- Li, Y.; Yang, C.; Zhang, L.; Yang, P. MicroRNA-210 induces endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis. Cell. Mol. Biol. Lett. 2017, 22, 3. [Google Scholar] [CrossRef] [Green Version]
- Raitoharju, E.; Lyytikäinen, L.-P.; Levula, M.; Oksala, N.; Mennander, A.; Tarkka, M.; Klopp, N.; Illig, T.; Kähönen, M.; Karhunen, P.J.; et al. miR-21, miR-210, miR-34a, and miR-146a/b are upregulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011, 219, 211–217. [Google Scholar] [CrossRef]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Treguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Merlet, E.; Atassi, F.; Motiani, R.K.; Mougenot, N.; Jacquet, A.; Nadaud, S.; Capiod, T.; Trebak, M.; Lompré, A.-M.; Marchand, A. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovasc. Res. 2013, 98, 458–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.-C.; Garmire, L.X.; Young, A.; Nguyen, P.; Trinh, A.; Subramaniam, S.; Wang, N.; Shyy, J.Y.; Li, Y.-S.; Chien, S. Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth. Proc. Natl. Acad. Sci. USA 2010, 107, 3234–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Friggeri, A.; Yang, Y.; Park, Y.-J.; Tsuruta, Y.; Abraham, E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. USA 2009, 106, 15819–15824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remus, E.W.; Lyle, A.N.; Weiss, D.; Landázuri, N.; Weber, M.; Searles, C.; Taylor, W.R. miR181a protects against angiotensin II-induced osteopontin expression in vascular smooth muscle cells. Atherosclerosis 2013, 228, 168–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Gong, Y.; Yuan, J.; Zhang, W.; Zhao, G.; Li, H.; Sun, A.; Zou, Y.; Ge, J. microRNA-181a represses ox-LDL-stimulated inflammatory response in dendritic cell by targeting c-Fos[S]. J. Lipid Res. 2012, 53, 2355–2363. [Google Scholar] [CrossRef]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Vera, M.P.; Registry, M.; Blackwell, T.S.; et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar] [CrossRef]
- Xu, J.; Hu, G.; Lu, M.; Xiong, Y.; Li, Q.; Chang, C.C.; Song, B.; Chang, T.-Y.; Li, B. MiR-9 reduces human acyl-coenzyme A:cholesterol acyltransferase-1 to decrease THP-1 macrophage-derived foam cell formation. Acta Biochim. et Biophys. Sin. 2013, 45, 953–962. [Google Scholar] [CrossRef] [Green Version]
- Magenta, A.; Cencioni, C.; Fasanaro, P.; Zaccagnini, G.; Greco, S.; Sarra-Ferraris, G.; Antonini, A.; Martelli, F.; Capogrossi, M.C. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011, 18, 1628–1639. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.-H.; Lee, M.-J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLOS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nature 2011, 13, 423–433. [Google Scholar] [CrossRef]
- McManus, D.D.; Ambros, V. Circulating MicroRNAs in cardiovascular disease. Circulation 2011, 124, 1908–1910. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Da-Silva, T.; Cruz, M.C.; Carrusca, C.; Ferreira, R.C.; Napoleão, P.; Carmo, M.M. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: A systematic review. Am. J. Cardiovasc. Dis. 2018, 8, 1–13. [Google Scholar]
- Li, T.; Cao, H.; Zhuang, J.; Wan, J.; Guan, M.; Yu, B.; Li, X.; Zhang, W. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin. Chim. Acta 2011, 412, 66–70. [Google Scholar] [CrossRef]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Röxe, T.; Müller-Ardogan, M.; et al. Circulating MicroRNAs in Patients with Coronary Artery Disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef]
- Wang, G.-K.; Zhu, J.-Q.; Zhang, J.-T.; Li, Q.; He, J.; Qin, Y.-W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Hear. J. 2010, 31, 659–666. [Google Scholar] [CrossRef]
- Meder, B.; Keller, A.; Vogel, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Just, S.; Borries, A.; Rudloff, J.; Leidinger, P.; et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 2011, 106, 13–23. [Google Scholar] [CrossRef]
- Eken, S.M.; Jin, H.; Chernogubova, E.; Li, Y.; Simon, N.; Sun, C.; Korzunowicz, G.; Busch, A.; Bäcklund, A.; Österholm, C.; et al. MicroRNA-210 Enhances Fibrous Cap Stability in Advanced Atherosclerotic Lesions. Circ. Res. 2017, 120, 633–644. [Google Scholar] [CrossRef]
- Cipollone, F.; Felicioni, L.; Sarzani, R.; Ucchino, S.; Spigonardo, F.; Mandolini, C.; Malatesta, S.; Bucci, M.; Mammarella, C.; Santovito, D.; et al. A Unique MicroRNA Signature Associated with Plaque Instability in Humans. Stroke 2011, 42, 2556–2563. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.S.; Kim, J.Y.; Lee, S.H.; Hwang, J.; Shin, J.W.; Lee, S.; Kim, J. Abstract WP541: Profiling of MicroRNA from Atherosclerotic Tissues and Validation of Their Significance from Peripheral Blood of Ischemic Stroke Patients Having Atherosclerosis. Stroke 2019, 50, AWP541. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, M.; Sanagawa, A.; Mori, C.; Ito, S.; Iwaki, S.; Satoh, H.; Fujii, S. Circulating microRNA-126 in patients with coronary artery disease: Correlation with LDL cholesterol. Thromb. J. 2012, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xuekelati, S.; Zhou, K.; Yan, Z.; Yang, X.; Inayat, A.; Wu, J.; Guo, X. Expression Profiles of Six Atherosclerosis-Associated microRNAs That Cluster in Patients with Hyperhomocysteinemia: A Clinical Study. DNA Cell Biol. 2018, 37, 189–198. [Google Scholar] [CrossRef]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; Van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011, 478, 404–407. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; He, S.; Wara, A.K.M.; Icli, B.; Shvartz, E.; Tesmenitsky, Y.; Belkin, N.; Li, D.; Blackwel, T.S.; Sukhova, G.K.; et al. Systemic Delivery of MicroRNA-181b Inhibits Nuclear Factor-κB Activation, Vascular Inflammation, and Atherosclerosis in Apolipoprotein E–Deficient Mice. Circ. Res. 2014, 114, 32–40. [Google Scholar] [CrossRef]
- Guo, F.; Tang, C.; Li, Y.; Liu, Y.; Lv, P.; Wang, W.; Mu, Y. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-κB signalling pathway. J. Cell. Mol. Med. 2018, 22, 5062–5075. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; A Sharp, P. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Wen, Z.; Zheng, S.; Zhou, C.; Yuan, W.; Wang, J.; Wang, T. Bone marrow mesenchymal stem cells for post-myocardial infarction cardiac repair: microRNAs as novel regulators. J. Cell. Mol. Med. 2002, 16, 657–671. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Cui, Y.; Yang, J.J.; Zhang, Z.G.; Chopp, M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. Br. J. Pharmacol. 2013, 33, 1711–1715. [Google Scholar] [CrossRef]
- Thomas, G.S.; Cromwell, W.C.; Ali, S.; Chin, W.; Flaim, J.D.; Davidson, M. Mipomersen, an Apolipoprotein B Synthesis Inhibitor, Reduces Atherogenic Lipoproteins in Patients with Severe Hypercholesterolemia at High Cardiovascular Risk: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Am. Coll. Cardiol. 2013, 62, 2178–2184. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Rooij, E.; Purcell, A.L.; Levin, A.A. Developing microRNA therapeutics. Circ. Res. 2012, 110, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjärn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to upregulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2007, 36, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Piras, B.A.; O’Connor, D.M.; French, B.A. Systemic Delivery of shRNA by AAV9 Provides Highly Efficient Knockdown of Ubiquitously Expressed GFP in Mouse Heart, but Not Liver. PLoS ONE 2013, 8, e75894. [Google Scholar] [CrossRef]


| miRNA | Target | Function | References |
|---|---|---|---|
| miR-27a/b | SEM6A | Regulates sprouting angiogenesis of endothelial cells | [128] |
| miR-126-5p | SetD5 Dlk1 ALCAM | Promotes endothelial cell proliferation and repair; regulates leucocyte adhesion and transmigration | [129,130] |
| miR-126 | G protein | Reduces the macrophage and apoptotic cell content of plaques leading to lesion reduction in size with a less inflammatory phenotype; governs vascular integrity and angiogenesis | [131] |
| miR-302a | ABCA1 | Regulates cellular cholesterol efflux in macrophages | [132] |
| miR-758 | ABCA1 | Regulates macrophage cholesterol efflux protecting cells from the excessive lipid accumulation | [133] |
| miR-342-5p | AKT1 | Enhances proinflammatory macrophage mediators, such as iNOS and IL-6, via suppression of miR-155 expression | [134] |
| miR-370 | Regulates lipid metabolism by mediating synthesis of cholesterol, fatty acid, and fatty acid β-oxidation; regulates miR-122 levels | [135] | |
| miR-10a | MAP3K7, β-TRC | Inhibits NF-κB activation | [136] |
| miR-663 | JunB Myosin light chain 9 | Regulates monocyte adhesion induced by oscillatory shear stress, therefore, it involved in oscillatory shear stress-induced cellular inflammation; regulates VSMC phenotypic switch and vascular neointimal formation | [137,138] |
| miR-210 | EFNA3 PDK1 | Linked to intraplaque angiogenesis and, possibly, to the formation of unstable plaques; regulates endothelial apoptosis | [139,140] |
| miR-34a | PNUTS | Regulates cardiac aging and contractile function | [141] |
| miR-424/322 | Calumenin STIM1 Cyclin D1 | Regulates proliferation, migration, and differentiation of VSMCs | [142] |
| miR-23b | E2F1 Rb | Inhibits endothelial cell proliferation | [143] |
| miR-29 | ADAMTS-7 | Mediates VSMC calcification | [117] |
| miR-147 | TLR2 TLR3 TLR4 | Negatively regulates the TLR-associated signaling pathway increasing inflammatory cytokine expression in macrophages, hence, prevents excessive inflammatory responses | [144] |
| miR-181a | c-Fos OPN | Diminishes the oxLDL-induced immune-inflammatory response by downgrading dendritic maturation of cell surface molecules, such as CD40 and CD83; regulates endothelial cell proliferation; protects against angiotensin II-induced osteopontin expression in VSMCs | [145,146] |
| miR-181b | NF-κB | Suppresses the endothelial inflammatory response | [147] |
| miR-9 | ACAT1 | Regulates cellular cholesterol homeostasis by decreasing the formation of foam cells from macrophages | [148] |
| miR-200c | ZEB1 | Upregulated upon oxidative stress, induces apoptosis and senescence of endothelial cells | [149] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Churov, A.; Summerhill, V.; Grechko, A.; Orekhova, V.; Orekhov, A. MicroRNAs as Potential Biomarkers in Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 5547. https://doi.org/10.3390/ijms20225547
Churov A, Summerhill V, Grechko A, Orekhova V, Orekhov A. MicroRNAs as Potential Biomarkers in Atherosclerosis. International Journal of Molecular Sciences. 2019; 20(22):5547. https://doi.org/10.3390/ijms20225547
Chicago/Turabian StyleChurov, Alexey, Volha Summerhill, Andrey Grechko, Varvara Orekhova, and Alexander Orekhov. 2019. "MicroRNAs as Potential Biomarkers in Atherosclerosis" International Journal of Molecular Sciences 20, no. 22: 5547. https://doi.org/10.3390/ijms20225547