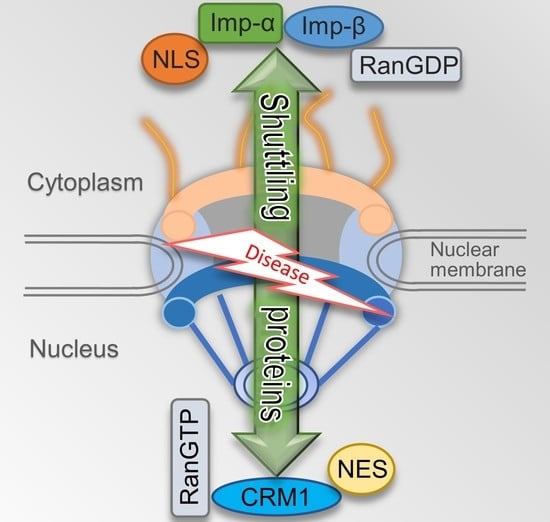
The Rules and Functions of Nucleocytoplasmic Shuttling Proteins
Abstract
:1. Introduction
2. The Mechanism of Shuttling Proteins
2.1. Structure and Function of the Nuclear Pore Complex (NPC)
2.2. Transport Signals for Nuclear Import and Export
2.2.1. Nuclear Localization Signals
2.2.2. Nuclear Export Signals
2.3. Transport Receptor Family
2.4. The Ran System
2.5. Nuclear Import of Proteins
2.6. Nuclear Export of Proteins
3. Shuttling Proteins and Diseases
3.1. Shuttling Proteins in Cancer
3.2. Shuttling Proteins in Neurodegenerative Diseases
3.3. Shuttling Proteins in Other Diseases
3.3.1. Shuttling Proteins in Acute Myeloid Leukemia
3.3.2. Shuttling Proteins in Osteoporosis
3.3.3. Shuttling Proteins in Fanconi Anemia
4. Perspective and Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T. The molecular architecture of the nuclear pore complex. Nature 2007, 450, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L. Localization of nucleus-specific protein as shown by transplantation experiments in Amoeba proteus. Exp. Cell Res. 1958, 15, 635–637. [Google Scholar] [CrossRef]
- Borer, R.; Lehner, C.; Eppenberger, H.; Nigg, E. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989, 56, 379–390. [Google Scholar] [CrossRef]
- Baranek, C.; Sock, E.; Wegner, M. The POU protein Oct-6 is a nucleocytoplasmic shuttling protein. Nucleic Acids Res. 2005, 33, 6277–6286. [Google Scholar] [CrossRef] [PubMed]
- Klibanov, S.A.; O’Hagan, H.M.; Ljungman, M. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J. Cell Sci. 2001, 114, 1867–1873. [Google Scholar] [PubMed]
- Stocco, D.M. StAR protein and the regulation of steroid hormone biosynthesis. Annu. Rev. Physiol. 2001, 63, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Dostie, J.; Ferraiuolo, M.; Pause, A.; Adam, S.A.; Sonenberg, N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000, 19, 3142–3156. [Google Scholar] [CrossRef] [PubMed]
- Shyu, A.B.; Wilkinson, M.F. The double lives of shuttling mRNA binding proteins. Cell 2000, 102, 135–138. [Google Scholar] [CrossRef]
- Schmidt-Zachmann, M.S.; Dargemont, C.; Kuhn, L.C.; Nigg, E.A. Nuclear export of proteins: The role of nuclear retention. Cell 1993, 74, 493–504. [Google Scholar] [CrossRef]
- Arenzana-Seisdedos, F.; Thompson, J.; Rodriguez, M.S.; Bachelerie, F.; Thomas, D.; Hay, R.T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol. Cell. Biol. 1995, 15, 2689–2696. [Google Scholar] [CrossRef] [PubMed]
- Mattaj, I.W.; Englmeier, L. Nucleocytoplasmic transport: The soluble phase. Annu. Rev. Biochem. 1998, 67, 265–306. [Google Scholar] [CrossRef] [PubMed]
- Fried, H.; Kutay, U. Nucleocytoplasmic transport: Taking an inventory. Cell. Mol. Life Sci. 2003, 60, 1659–1688. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Forster, F.; Ecke, M.; Plitzko, J.M.; Melchior, F.; Gerisch, G.; Baumeister, W.; Medalia, O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 2004, 306, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Stoffler, D.; Fahrenkrog, B.; Aebi, U. The nuclear pore complex: From molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 1999, 11, 391–401. [Google Scholar] [CrossRef]
- Devos, D.; Dokudovskaya, S.; Williams, R.; Alber, F.; Eswar, N.; Chait, B.T.; Rout, M.P.; Sali, A. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2006, 103, 2172–2177. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Dokudovskaya, S.; Alber, F.; Williams, R.; Chait, B.T.; Sali, A.; Rout, M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004, 2, e380. [Google Scholar] [CrossRef] [PubMed]
- Strawn, L.A.; Shen, T.; Shulga, N.; Goldfarb, D.S.; Wente, S.R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 2004, 6, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.B.; Gorlich, D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem. Sci. 2016, 41, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Kim, E.R.; Ovchinnikov, L.P. Nucleocytoplasmic transport of proteins. Biochemistry 2007, 72, 1439–1457. [Google Scholar] [CrossRef] [PubMed]
- Dingwall, C.; Sharnick, S.V.; Laskey, R.A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1982, 30, 449–458. [Google Scholar] [CrossRef]
- Dingwall, C.; Laskey, R.A. Nuclear targeting sequences—A consensus? Trends Biochem. Sci. 1991, 16, 478–481. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, H.H.; Fu, J.; Wang, X.F.; Ren, Y.B.; Dong, L.W.; Tang, S.H.; Liu, S.Q.; Wu, M.C.; Wang, H.Y. Oncoprotein p28 GANK binds to RelA and retains NF-κB in the cytoplasm through nuclear export. Cell Res. 2007, 17, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, H.; Xu, X.; Zhou, H.; Li, X.; Peng, C.; Shen, S.; Xiong, W.; Ma, J.; Zeng, Z.; et al. Identification of nuclear localization signal that governs nuclear import of BRD7 and its essential roles in inhibiting cell cycle progression. J. Cell. Biochem. 2006, 98, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; McLane, L.M.; Mills, R.E.; Devine, S.E.; Corbett, A.H. Expanding the definition of the classical bipartite nuclear localization signal. Traffic 2010, 11, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Farmer, A.; Chook, Y.M. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr. Opin. Struct. Biol. 2010, 20, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin β2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Antoine, M.; Reimers, K.; Wirz, W.; Gressner, A.M.; Muller, R.; Kiefer, P. Identification of an unconventional nuclear localization signal in human ribosomal protein S2. Biochem. Biophys. Res. Commun. 2005, 335, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattajt, L.W.; Lührmann, R. The HIV-1 Rev Activation Domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef]
- Wen, W.; Meinkoth, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Molgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Paraskeva, E.; Izaurralde, E.; Bischoff, F.R.; Huber, J.; Kutay, U.; Hartmann, E.; Luhrmann, R.; Gorlich, D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 1999, 145, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Taagepera, S.; McDonald, D.; Loeb, J.E.; Whitaker, L.L.; McElroy, A.K.; Wang, J.Y.; Hope, T.J. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc. Natl. Acad. Sci. USA 1998, 95, 7457–7462. [Google Scholar] [CrossRef] [PubMed]
- Ossareh-Nazari, B.; Bachelerie, F.; Dargemont, C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 1997, 278, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Haasen, D.; Kohler, C.; Neuhaus, G.; Merkle, T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999, 20, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Conti, E.; Izaurralde, E. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 2001, 13, 310–319. [Google Scholar] [CrossRef]
- Kohler, M.; Fiebeler, A.; Hartwig, M.; Thiel, S.; Prehn, S.; Kettritz, R.; Luft, F.C.; Hartmann, E. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 2002, 12, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D.S.; Corbett, A.H.; Mason, D.A.; Harreman, M.T.; Adam, S.A. Importin α: A multipurpose nuclear-transport receptor. Curr. Opin. Cell Biol. 2004, 14, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Moroianu, J.; Blobel, G.; Radu, A. The binding site of karyopherin α for karyopherin β overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. USA 1996, 93, 6572–6576. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, L.F.; Paschal, B.M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 2005, 6, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Conti, E.; Müller, C.W.; Stewart, M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006, 16, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Weis, K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell 2003, 112, 441–451. [Google Scholar] [CrossRef]
- Ohno, M.; Segref, A.; Bachi, A.; Wilm, M.; Mattaj, I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000, 101, 187–198. [Google Scholar] [CrossRef]
- Izaurralde, E.; Lewis, J.; Gamberi, C.; Jarmolowski, A.; McGuigan, C.; Mattaj, I.W. A cap-binding protein complex mediating U snRNA export. Nature 1995, 376, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Arts, G.J.; Kuersten, S.; Romby, P.; Ehresmann, B.; Mattaj, I.W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998, 17, 7430–7441. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004, 14, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, A.; Kurisaki, K.; Kowanetz, M.; Sugino, H.; Yoneda, Y.; Heldin, C.H.; Moustakas, A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell. Biol. 2006, 26, 1318–1332. [Google Scholar] [CrossRef] [PubMed]
- Kutay, U.; Bischoff, F.R.; Kostka, S.; Kraft, R.; Gorlich, D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef]
- Kimura, M.; Morinaka, Y.; Imai, K.; Kose, S.; Horton, P.; Imamoto, N. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. Elife 2017, 6, e21184. [Google Scholar] [CrossRef] [PubMed]
- Mackmull, M.T.; Klaus, B.; Heinze, I.; Chokkalingam, M.; Beyer, A.; Russell, R.B.; Ori, A.; Beck, M. Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol. 2017, 13, 962. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Hayashi, N.; Noguchi, E.; Nishimoto, T. Putative GTPase Gtr1p genetically interacts with the RanGTPase cycle in Saccharomyces cerevisiae. J. Cell Sci. 1996, 109, 2311–2318. [Google Scholar] [PubMed]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Koepp, D.M.; Silver, P.A. A GTPase controlling nuclear trafficking: Running the right way or walking RANdomly? Cell 1996, 87, 1–4. [Google Scholar] [CrossRef]
- Cook, A.; Bono, F.; Jinek, M.; Conti, E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007, 76, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Pante, N.; Kutay, U.; Aebi, U.; Bischoff, F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996, 15, 5584–5594. [Google Scholar] [PubMed]
- Moroianu, J.; Blobel, G.; Radu, A. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing α from an overlapping binding site on β. Proc. Natl. Acad. Sci. USA 1996, 93, 7059–7062. [Google Scholar] [CrossRef] [PubMed]
- Gasiorowski, J.Z.; Dean, D.A. Mechanisms of nuclear transport and interventions. Adv. Drug Deliv. Rev. 2003, 55, 703–716. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Arnaoutov, A.; Azuma, Y.; Ribbeck, K.; Joseph, J.; Boyarchuk, Y.; Karpova, T.; McNally, J.; Dasso, M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005, 7, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Englmeier, L.; Fornerod, M.; Bischoff, F.R.; Petosa, C.; Mattaj, I.W.; Kutay, U. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep. 2001, 2, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Biswas, A.; Chook, Y.M. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat. Struct. Mol. Biol. 2009, 16, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Surget, S.; Khoury, M.P.; Bourdon, J.C. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2013, 7, 57–68. [Google Scholar]
- Moll, U.M.; LaQuaglia, M.; Benard, J.; Riou, G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc. Natl. Acad. Sci. USA 1995, 92, 4407–4411. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Li, A.G.; Sheppard, H.M.; Liu, X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: A role for TAF1 in cell G1 progression. Mol. Cell 2004, 13, 867–878. [Google Scholar] [CrossRef]
- Cai, X.; Liu, X. Inhibition of Thr-55 phosphorylation restores p53 nuclear localization and sensitizes cancer cells to DNA damage. Proc. Natl. Acad. Sci. USA 2008, 105, 16958–16963. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, R.J.; Ironside, J.A.D. Squamous cell carcinomas of the head and neck. Br. Med. J. 2002, 325, 822–827. [Google Scholar] [CrossRef] [Green Version]
- Mandic, R.; Schamberger, C.J.; Muller, J.F.; Geyer, M.; Zhu, L.; Carey, T.E.; Grenman, R.; Dunne, A.A.; Werner, J.A. Reduced cisplatin sensitivity of head and neck squamous cell carcinoma cell lines correlates with mutations affecting the COOH-terminal nuclear localization signal of p53. Clin. Cancer Res. 2005, 11, 6845–6852. [Google Scholar] [CrossRef] [PubMed]
- El-Tanani, M.; Dakir, E.H.; Raynor, B.; Morgan, R. Mechanisms of Nuclear Export in Cancer and Resistance to Chemotherapy. Cancers 2016, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Altieri, D.C. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 2006, 18, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Lens, S.M.; Wolthuis, R.M.; Klompmaker, R.; Kauw, J.; Agami, R.; Brummelkamp, T.; Kops, G.; Medema, R.H. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003, 22, 2934–2947. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Kramer, O.H.; Knosel, T.; Engels, K.; Rodel, F.; Kovacs, A.F.; Dietmaier, W.; Klein-Hitpass, L.; Habtemichael, N.; Schweitzer, A.; et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 2007, 21, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Unruhe, B.; Karczewski, S.; Hecht, R.; Fetz, V.; Bier, C.; Friedl, S.; Wollenberg, B.; Pries, R.; Habtemichael, N.; et al. Functional characterization of novel mutations affecting survivin (BIRC5)-mediated therapy resistance in head and neck cancer patients. Hum. Mutat. 2013, 34, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Bennit, H.F.; Wall, N.R. The emerging role of exosomes in survivin secretion. Histol. Histopathol. 2015, 30, 43–50. [Google Scholar] [PubMed]
- Fong, L.W.R.; Yang, D.C.; Chen, C.H. Myristoylated alanine-rich C kinase substrate (MARCKS): A multirole signaling protein in cancers. Cancer Metastasis Rev. 2017, 36, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Disatnik, M.-H.; Boutet, S.C.; Pacio, W.; Chan, A.Y.; Ross, L.B.; Lee, C.H.; Rando, T.A. The bi-directional translocation of MARCKS between membrane and cytosol regulates integrin-mediated muscle cell spreading. J. Cell Sci. 2004, 117, 4469–4479. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Rotenberg, S.A. PhosphoMARCKS drives motility of mouse melanoma cells. Cell. Signal. 2010, 22, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-H.; Statt, S.; Chiu, C.-L.; Thai, P.; Arif, M.; Adler, K.B.; Wu, R. Targeting Myristoylated Alanine-Rich C Kinase Substrate Phosphorylation Site Domain in Lung Cancer. Mechanisms and Therapeutic Implications. Am. J. Respir. Crit. Care Med. 2014, 190, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Rohrbach, T.D.; Shah, N.; Jackson, W.P.; Feeney, E.V.; Scanlon, S.; Gish, R.; Khodadadi, R.; Hyde, S.O.; Hicks, P.H.; Anderson, J.C.; et al. The Effector Domain of MARCKS Is a Nuclear Localization Signal that Regulates Cellular PIP2 Levels and Nuclear PIP2 Localization. PLoS ONE 2015, 10, e0140870. [Google Scholar] [CrossRef] [PubMed]
- Bugler, B.; Caizergues-Ferrer, M.; Bouche, G.; Bourbon, H.; Amalric, F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur. J. Biochem. 1982, 128, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Semenkovich, C.F.; Ostlund, R.E.; Olson, M.O.J.; Yang, J.W. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry 1990, 29, 9708–9713. [Google Scholar] [CrossRef] [PubMed]
- Galzio, R.; Rosati, F.; Benedetti, E.; Cristiano, L.; Aldi, S.; Mei, S.; D’Angelo, B.; Gentile, R.; Laurenti, G.; Cifone, M.G.; et al. Glycosilated nucleolin as marker for human gliomas. J. Cell. Biochem. 2012, 113, 571–579. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, D.; Destouches, D.; Lengagne, R.; Krust, B.; Hamma-Kourbali, Y.; Garcette, M.; Niro, S.; Kato, M.; Briand, J.P.; Courty, J.; et al. Targeting surface nucleolin with a multivalent pseudopeptide delays development of spontaneous melanoma in RET transgenic mice. BMC Cancer 2010, 10, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Richmond, T.; Piovan, C.; Sheetz, T.; Zanesi, N.; Troise, F.; James, C.; Wernicke, D.; Nyei, F.; Gordon, T.J.; et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 9418–9423. [Google Scholar] [CrossRef] [PubMed]
- Beal, M.F. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann. Neurol. 1995, 38, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.Y.; Goldberg, A.L. Cellular Defenses against Unfolded Proteins: A Cell Biologist Thinks about Neurodegenerative Diseases. Neuron 2001, 29, 15–32. [Google Scholar] [CrossRef]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz, S.; Cleveland, D.W. Disrupted nuclear import/export in neurodegeneration. Science 2016, 351, 125–126. [Google Scholar] [CrossRef] [PubMed]
- Woerner, A.C.; Frottin, F.; Hornburg, D.; Feng, L.R.; Meissner, F.; Patra, M.; Tatzelt, J.; Mann, M.; Winklhofer, K.F.; Hartl, F.U.; et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016, 351, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, O.; Zhu, X.; Lee, H.G.; Raina, A.; Obrenovich, M.E.; Bowser, R.; Ghanbari, H.A.; Castellani, R.J.; Perry, G.; Smith, M.A. Ectopic localization of phosphorylated histone H3 in Alzheimer’s disease: A mitotic catastrophe? Acta Neuropathol. 2003, 105, 524–528. [Google Scholar] [PubMed]
- Iqbal, K.; Tellez-Nagel, I.; Grundke-Iqbal, I. Protein abnormalities in Huntington’s chorea. Brain Res. 1974, 76, 178–184. [Google Scholar] [CrossRef]
- Ramsey, C.P.; Glass, C.A.; Montgomery, M.B.; Lindl, K.A.; Ritson, G.P.; Chia, L.A.; Hamilton, R.L.; Chu, C.T.; Jordan-Sciutto, K.L. Expression of Nrf2 in Neurodegenerative Diseases. J. Neuropathol. Exp. Neurol. 2007, 66, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Ueda, M.; Miyamoto, Y.; Yoneda, Y.; Perry, G.; Smith, M.A.; Zhu, X. Aberrant localization of importin α1 in hippocampal neurons in Alzheimer disease. Brain Res. 2006, 1124, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Ayala, Y.M.; Zago, P.; D’Ambrogio, A.; Xu, Y.-F.; Petrucelli, L.; Buratti, E.; Baralle, F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008, 121, 3778–3785. [Google Scholar] [CrossRef] [PubMed]
- Winton, M.J.; Igaz, L.M.; Wong, M.M.; Kwong, L.K.; Trojanowski, J.Q.; Lee, V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008, 283, 13302–13309. [Google Scholar] [CrossRef] [PubMed]
- Bosco, D.A.; Lemay, N.; Ko, H.K.; Zhou, H.; Burke, C.; Kwiatkowski, J.T.J.; Sapp, P.; McKenna-Yasek, D.; Brown, J.R.H.; Hayward, L.J. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 2010, 19, 4160–4175. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Taylor, J.P. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 2017, 96, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, G.; Dell’Oro, M.G.; Mecucci, C.; Giarin, E.; Masetti, R.; Rossi, V.; Locatelli, F.; Martelli, M.F.; Basso, G.; Pession, A.; et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood 2005, 106, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- Mariano, A.R.; Colombo, E.; Luzi, L.; Martinelli, P.; Volorio, S.; Bernard, L.; Meani, N.; Bergomas, R.; Alcalay, M.; Pelicci, P.G. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene 2006, 25, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- Morinobu, M.; Nakamoto, T.; Hino, K.; Tsuji, K.; Shen, Z.J.; Nakashima, K.; Nifuji, A.; Yamamoto, H.; Hirai, H.; Noda, M. The nucleocytoplasmic shuttling protein CIZ reduces adult bone mass by inhibiting bone morphogenetic protein-induced bone formation. J. Exp. Med. 2005, 201, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Hino, K.; Nakamoto, T.; Nifuji, A.; Morinobu, M.; Yamamoto, H.; Ezura, Y.; Noda, M. Deficiency of CIZ, a nucleocytoplasmic shuttling protein, prevents unloading-induced bone loss through the enhancement of osteoblastic bone formation in vivo. Bone 2007, 40, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Lin, X.; Xu, P.; Zhang, Z.; Chen, Y.; Wang, C.; Han, J.; Zhao, B.; Xiao, M.; Feng, X.H. Nuclear Export of Smads by RanBP3L Regulates Bone Morphogenetic Protein Signaling and Mesenchymal Stem Cell Differentiation. Mol. Cell. Biol. 2015, 35, 1700–1711. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, R.A.; Rego, M.A.; Azzinaro, P.A.; Mauro, M.; Howlett, N.G. Coordinate Nuclear Targeting of the FANCD2 and FANCI Proteins via a FANCD2 Nuclear Localization Signal. PLoS ONE 2013, 8, e81387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.G.; Dawson, J.; Sullivan, D.M. Nuclear export of proteins and drug resistance in cancer. Biochem. Pharmacol. 2012, 83, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Liang, C.; Li, F.; Wang, L.; Wu, X.; Lu, A.; Xiao, G.; Zhang, G. The Rules and Functions of Nucleocytoplasmic Shuttling Proteins. Int. J. Mol. Sci. 2018, 19, 1445. https://doi.org/10.3390/ijms19051445
Fu X, Liang C, Li F, Wang L, Wu X, Lu A, Xiao G, Zhang G. The Rules and Functions of Nucleocytoplasmic Shuttling Proteins. International Journal of Molecular Sciences. 2018; 19(5):1445. https://doi.org/10.3390/ijms19051445
Chicago/Turabian StyleFu, Xuekun, Chao Liang, Fangfei Li, Luyao Wang, Xiaoqiu Wu, Aiping Lu, Guozhi Xiao, and Ge Zhang. 2018. "The Rules and Functions of Nucleocytoplasmic Shuttling Proteins" International Journal of Molecular Sciences 19, no. 5: 1445. https://doi.org/10.3390/ijms19051445