- Department of Pharmacology & Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
Ovarian cancer is one of the top gynecological malignancies that cause deaths among females in the United States. At the molecular level, significant progress has been made in our understanding of ovarian cancer development and progression. MicroRNAs (miRNAs) are short, single-stranded, highly conserved non-coding RNA molecules (19–25 nucleotides) that negatively regulate target genes post-transcriptionally. Over the last two decades, mounting evidence has demonstrated the aberrant expression of miRNAs in different human malignancies, including ovarian carcinomas. Deregulated miRNAs can have profound impacts on various cancer hallmarks by repressing tumor suppressor genes. This review will discuss up-to-date knowledge of how the aberrant expression of miRNAs and their targeted genes drives ovarian cancer initiation, proliferation, survival, and resistance to chemotherapies. Understanding the mechanisms by which these miRNAs affect these hallmarks should allow the development of novel therapeutic strategies to treat these lethal malignancies.
Introduction
Ovarian cancer is the eighth leading cause of cancer-related deaths among females worldwide, with an estimated 295,414 new cases and 184,799 deaths as of 2018 (1). Even though the 5-year relative survival rate at stages I–II ranges between 75 and 92%, in the western world, for instance, the rate remains <30% for patients presenting at the clinic with advanced peritoneal dissemination and massive ascites (2). One of the main reasons for the high mortality rates is the lack of an efficient and sensitive method that can detect ovarian cancer at early stages. Current diagnostic approaches, including serum levels of CA125, pelvic examination, and transvaginal ultrasonography, have failed to detect this disease at the early stages (3).
Approximately 90% of primary malignant ovarian tumors arise from the ovarian surface epithelium (OSE). Morphologically, epithelial ovarian carcinomas are divided into four major types (serous, endometrioid, clear cell, and mucinous) (4). The heterogeneity of ovarian cancer is suggested to account for the high mortality rates caused by this disease (5). Ovarian carcinomas are also classified into low-grade and high-grade cancers based on their expression profiles of signature proteins or their characterized mutations, which define their aggressiveness and response to chemotherapies. At the molecular level, low-grade ovarian tumors characterized by several signature mutations, including PTEN, PIK3CA KRAS, BRAF, ERBB2, and ARID1A, whereas the more aggressive high-grade tumors are associated with TP53 mutations (6). In addition to the importance of these protein-coding genes, advances in genetics revealed that the mammalian genome transcribes thousands of short non-protein-coding RNAs that regulate various biological processes, among which are microRNAs (miRNAs). Extensive analysis of more than 1493 small RNA deep sequencing datasets estimated that the human genome transcribes about 1917 miRNA precursor sequences, generating 2654 mature miRNA sequences (7). MiRNAs are short, single-stranded, highly conserved non-coding RNA molecules (19–25 nucleotides) that negatively regulate the targeted protein-coding genes post-transcriptionally through binding with their 3′-UTR (untranslated region). Such binding facilitates the degradation of these genes' mRNAs or blocks their translation (8). The synthesis and processing of miRNAs are illustrated elsewhere (9). Briefly, miRNAs are first transcribed in the nucleus by the RNA polymerase II into long, double-stranded precursors called primary miRNA transcripts (pri-miRNAs), which are then processed by the nuclear RNase III microprocessor complex Drosha and DGCR8 to produce 60–70-nucleotide RNA hairpin-like intermediates called premature miRNAs (pre-miRNAs). The pre-miRNAs are then translocated primely to the cytoplasm by a RNA.GTP-bound protein, exportin-5, where the latter is cleaved by another RNase III enzyme, the Dicer complex, into 19–25-nucleotide RNAs (mature miRNAs). Mature miRNAs are then incorporated into a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC), which, in collaboration with the Argonaute (AGO) protein, directs the miRNA complex to the targeted mRNA and facilitates its binding through perfect or imperfect base pairing. Since the miRNA binding to the 3′-UTR of the targeted mRNA does not require perfect complementarity, in addition to its ability to recognize strands as short as 2–8 nucleotides complementary to their 5′-seeding region, it has been estimated that one miRNA is capable of regulating the expression of several 100 genes and that one mRNA is regulated by multiple miRNAs.
Recent bioinformatic analysis has predicted that approximately two-thirds of all human genes are regulated by more than 1,000 miRNAs (10), which has prompted scientists in the last decade to investigate the roles of these miRNAs in many diseases, including cancer. Emerging evidence suggests that deregulated miRNAs play important regulatory roles in the initiation, progression, and dissemination of different types of cancer (11). It has become well-established that miRNAs can either be upregulated or downregulated in various human cancers, including ovarian cancer. Oncogenesis can be initiated either by the overexpression of oncogenic miRNAs, which downregulate tumor suppressor genes, or by the loss of specific tumor-suppressive miRNAs, which negatively regulate oncogenes (12). Since the first report establishing a connection between miRNAs and cancer (13), the importance of aberrant miRNA expression in different malignancies has been firmly established (47,009 PubMed hits in February 2020). Of these, 1,714 PubMed hits report diagnostic, prognostic, and therapeutic implications of miRNAs in ovarian cancer. This review summarizes the mechanisms by which miRNAs drive the transformation and tumorigenesis of different types of ovarian carcinomas. This work provides a deeper understanding of how miRNAs and their respective, experimentally verified target genes control different aspects of ovarian cancer pathogenesis.
Aberrant Expression of miRNAs: Potential Diagnostic and Prognostic Biomarkers of Ovarian Cancer
miRNA Expression Profiles in Ovarian Cancer Tissues
A substantial number of studies have investigated the differential expression profiles of miRNAs in the serum, plasma, exosomes in serum, ascites fluids, and tissues of ovarian cancer patients and normal counterparts to identify potential diagnostic and prognostic biomarkers. One of the first reports investigating the miRNA expression profiles in tissues obtained from ovarian cancer patients and normal subjects revealed signature expression patterns between the two groups. Among the commonly upregulated miRNAs regardless of the morphological histotypes reported in this study were miR-141and miR-200a-c, whereas miR-125b, miR-140, miR-145, and miR-199a were among the downregulated miRNAs in ovarian cancer tissues. This study also reported distinctive expression profiles of miRNAs within ovarian cancer tissues with different histopathologic features (serous, endometrioid, clear cell, and mucinous). For example, when compared with serous, endometrioid histotype seems to significantly upregulate miR-212 and miR-302b*, and downregulate miR-222 (14). The Lower number of samples of clear cell and mucinous carcinoma subtypes makes it harder to draw solid conclusions or safely distinguish between expression profiles. In another study, Zhang et al. have also assessed the alterations in the expression levels of miRNAs and their association with the malignant transformation of the OSE, the origin of EOC (15). This study compared the mature miRNA expression profiles of 18 EOC cell lines and four immortalized non-malignant human ovarian surface epithelium (IOSE) primary cell lines using multiple comprehensive miRNome techniques. Their results showed that 31 miRNAs were downregulated in the EOC lines compared with the IOSE cell lines, including the tumor suppressor miRNAs lethal-7 (let-7d) (16) and miR-127 (17). Analysis of primary human ovarian cancer specimens revealed that late-stage tumors significantly downregulated tumor suppressors miRNAs, miR-15a, miR-34a, and miR-34b, when compared with early-stage specimens. In an attempt to discriminate between primary and recurrent ovarian cancers, Laios et al. utilized TaqMan RT-PCR assay to quantify the miRNA expression profiles of primary and recurrent serous papillary adenocarcinomas. Their study revealed significant up and downregulation of miR-223 and miR-9, respectively, in recurrent tumors relative to the primary ones (18). Kaplan-Meier analysis of 20 patients with serous ovarian carcinomas revealed that higher expression of miR-18a, miR-93, and miR-200 family members (miR-141, miR-429 miR-200a-c), and lower expression of let-7b, and miR-199a were significantly correlated with decreased progression-free survival (PFS) and overall survival (OS) (19). The similarity between identified miRNAs in this study and the dysregulated miRNAs reported by Iorio et al. (14) further insinuate the importance of miRNAs as prognostic biomarkers. A few years later, Calura et al. confirmed the results demonstrated by previous studies when they analyzed the miRNA profiles for each EOC histotype at early stages to identify signature biomarkers for different ovarian carcinoma subtypes. This study demonstrated histotype-specific miRNA signatures for clear cell and mucinous ovarian cancer tissues. The clear cell histotype was characterized by high expression of miR-30a-5p and miR-30a-3p, whereas miR-192 and miR-194 were upregulated in the mucinous histotype (20).
A recent study examined the differentially expressed miRNAs in high-grade serous ovarian carcinoma (HGSC), clear cell ovarian carcinoma (CCC) and OSE. Both malignant subtypes mostly overexpressed miR-200 family members and miR-182-5p, whereas miR-383 was significantly underexpressed relative to OSE expressions. Higher expression of miR-509-3-5p, miR-509-5p, miR-509-3p, and miR-510 distinguished CCC from HGSC. Despite the relatively small number of subjects, higher miR-200c-3p expression was associated with poor PFS and OS in HGSC patients (21). More subtypes-oriented research is needed with a larger number of participants to validate signature miRNA biomarkers for the diagnosis and prognosis of ovarian cancer patients. The increasing number of reports that miRNAs expression profiles could be influenced by ethnicity and genetic backgrounds (22–24), urges for considering such a factor in the future studies aiming for identifying miRNA-based biomarkers for ovarian carcinomas. In addition to the two reasons pointed out above, the discrepancies of reported miRNA signature profiles in the tissues of ovarian cancer patients can be attributed to the heterogeneity of dissected tissue sample, the way these samples were processed, the number of miRNAs analyzed, the molecular assays used to detect these miRNAs, and the lack of standard methods for normalization (25). A more consistent, non-invasive, and less biased source of sampling is likely to produce more robust, reliable, and unified results that could help in the diagnosis, prognosis, and treatment of ovarian cancer patients.
Circulatory miRNA Expression Profiles
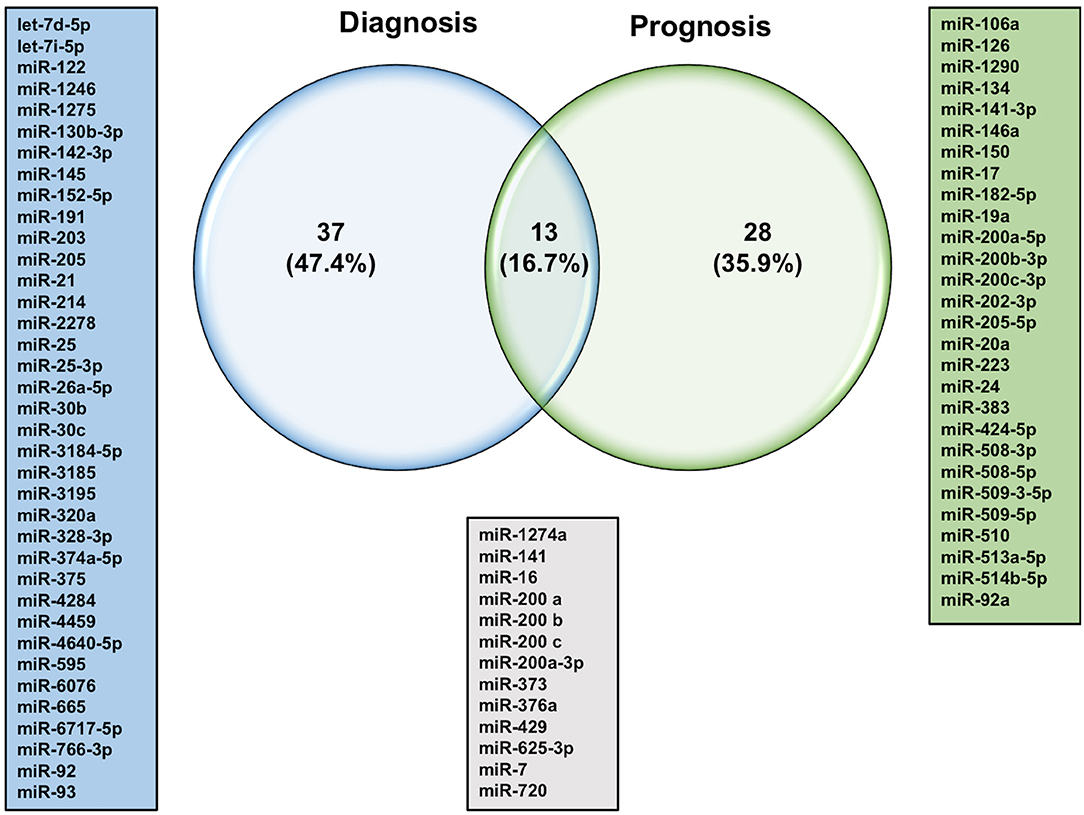
The quest for finding more reliable and distinctive diagnostic biomarkers for ovarian cancer, especially in the early stages, has been a priority of the scientific community for years (12). The differential expression of circulatory miRNAs has long been a more appealing approach for identifying clinical biomarkers for several reasons. miRNAs are found in the plasma and serum encapsulated in extracellular vesicles or bound with special lipid proteins, which make them resistant to RNase digestions (26). Several lines of evidence reported that these miRNAs have faster biogenesis and activation rates, and longer half-lives relative to mRNA and proteins, which make them suitable for detection at early stages (diagnostic), during progression or treatment selection (prognostic) (27, 28). One of the pitfalls of using single circulatory miRNA is the lack of specificity. For instance, several miRNAs such as miR-21-5p, miR-155-5p, and miR-210-3p have shown different expression patterns in different types of cancers, including lung, colorectal, and breast cancer (29). Therefore, the is an urgent need to employ advances in cancer genetics to identify specific miRNA signature patterns instead of stand-alone miRNA for the clinical diagnosis of ovarian cancer. Yokoi et al. have developed a miRNA-based detection method using comprehensive expression profiles of 10 serum-isolated miRNAs specific to ovarian tumors and helped discriminate ovarian tumors from other types of solid tumors and non-cancer cells (miR-320a, miR-665, miR-3184-5p, miR-6717-5p, miR-4459, miR-6076, miR-3195, miR-1275, miR-3185, and miR-4640-5p) (30). More specifically, a recent study identified miR-508-3p as a master regulator of the mesenchymal subtype of ovarian cancer and a promising prognostic biomarker that could help to diagnose this lethal disease in its early stages (31). Most of the studies that have investigated the expression profiles of miRNAs in the circulation of ovarian cancer patients are summarized in Table 1. Only studies that used samples from more than 20 ovarian cancer patients were included in this table. Some of these miRNAs showed consistent expression patterns in the circulation of ovarian cancer patients. For example, miR-25 was reported by two independent studies to be downregulated in the serum samples of ovarian cancer patients (40, 41). Pathway enrichment analysis revealed that miR-25-3p targets the oncogenic Toll-like receptor-4 (TLR-4)/MD88 pathway, which could result in overexpression of the latter (40). On the other hand, the vast majority of studies report significant upregulation of the miR-200 family members in the circulation of ovarian cancer patients except for one study that showed only miR-200c family member to be upregulated (47). The variability in sample processing and quantification techniques utilized in this study might count for undetectable levels of other family members, including miR-141, miR-200a, and miR-200b. This study differs from others that showed upregulated miR-200 family in two little details, the lower number of qRT-PCR cycles and the absence of the cDNA preamplification step. In addition, results from Table 1 were used to determine which miRNAs could serve as diagnostic and/or prognostic biomarkers. As shown in Figure 1, of the 50 dysregulated miRNAs that are reported here in ovarian cancer patients relative to healthy individuals, only 13 miRNAs (16.7%) demonstrated the ability to serve as diagnostic and prognostic biomarkers in ovarian cancer patients, among which are the miR-200 family members. Further extensive studies with a higher number of subjects are needed to investigate the expression patterns of those 13 miRNAs at different stages of the disease, their ability to distinguish different ovarian cancer subtypes, and their utilization as predictors of the disease outcomes and biomarkers for disease relapse.
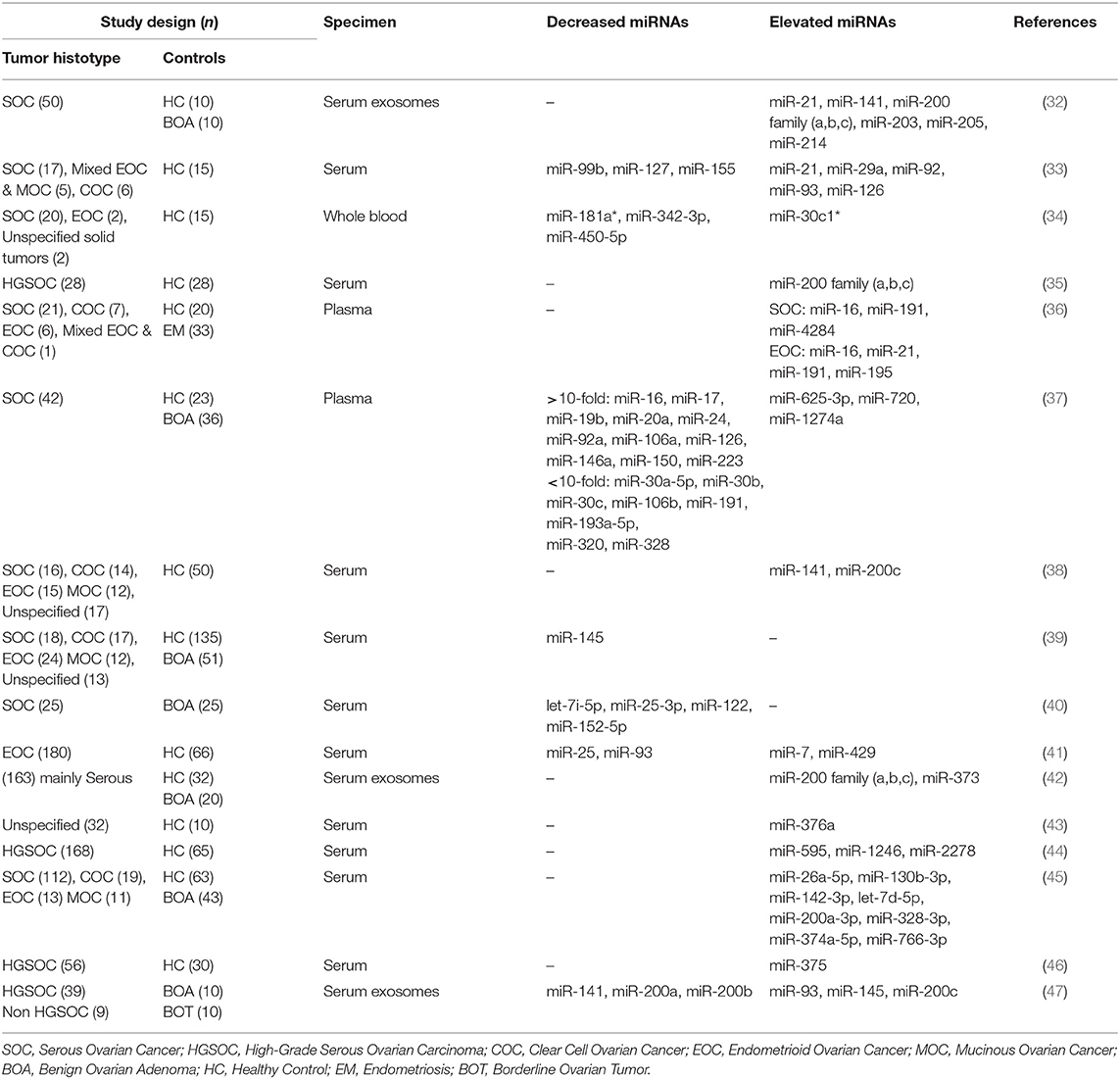
Table 1. MicroRNA expression profiles in different ovarian cancer subtypes in relation to normal controls.
MicroRNAs Regulate the Initiation of Ovarian Carcinomas
The initiation of cancer is a complex multi-step process that requires inactivation of tumor suppressor genes and the activation of oncogenes. The regulatory roles of miRNAs in the initiation of ovarian cancer comprise a rich area of research that has yet to be further explored. Substantial evidence has linked dysregulation in miRNAs expression with the early stages of ovarian cancer transformation. In humans, for example, the consensus let-7 family of tumor suppressor miRNAs has been suggested by numerous studies to be downregulated in several types of cancer, including breast, lung, stomach, colon, and ovarian cancer (48, 49). Among the reported target genes of the let-7 family are the early embryonic genes HMGA2 and IMP-1 (50) as well as oncogenic NRAS (16). The embryonic high mobility group AT-hook 2 (HMGA2), a direct target of let-7 (51–53), is a non-histone, nuclear-binding, oncofetal protein that has been shown to be overexpressed in embryonic tissue and many malignancies, including high-grade ovarian carcinomas (54, 55). In addition, HMGA2 functions as a crucial regulator of cell growth and differentiation (56). Shell et al. have provided the first direct association in human cancer between the downregulation of let-7 and the upregulation of HMGA2. Their study revealed that ovarian cancer patients with high HMGA2 and low let-7 expression in their cancer tissues had lower survival rates than patients with low HMGA2 and high let-7 expression (57). In another study, Wu et al. reported that overexpressing HMGA2 initiated the malignant transformation of OSE cell lines in vitro and in vivo and induced more aggressive tumor cell growth (55). Taken together, this evidence suggests that the downregulation of the let-7 family of miRNAs may serve a critical function in the malignant transformation of OSE into EOC. However, further loss and gain of function experiments are needed to demonstrate the direct impact of the tumor suppressor let-7 on the initiation and malignant transformation of ovarian cancer.
Previous studies have reported that ovarian cancer tissues had significantly lower miR-199a expression profiles relative to normal ovarian cells, which has been associated with poor PFS and OS in patients with serous ovarian carcinoma (14, 19). CD44+/CD177+ cells are ovarian cancer-initiating stem cells (CIC), which are known to be highly proliferative and resistant to chemotherapeutics, with low differentiation capabilities (58–60). Findings from a recent study by Cheng et al. revealed that miR-199a targets CD44 in these cells resulting in significant suppression of cell cycle, proliferation capacity, and invasiveness in vitro and tumor growth in vivo (61). These data suggest that the deregulation of miR-199a expression may serve as a prerequisite for the initiation of ovarian cancer.
During the differentiation process, tumor-initiating stem cells lose their stemness and acquire the characteristics of fast-growing, chemoresistant mature cancer cells. For instance, twist-related protein 1 (Twist1)-mediated upregulation of miR-199a/214 has been shown to be essential for the transformation of ovarian cancer stem cells (Type I/CD44+) into mature, fast-dividing cancer cells (Type II/CD44−) (62). At the molecular level, the upregulation of miR-214 and miR-199a resulted in a reduction of the tumor suppressors PTEN and IKKβ, which led to the activation of the AKT and NFκB pathways, respectively. All these independent findings further suggest that miRNAs can serve as vital regulators of the initiation of ovarian cancer. Further studies are needed to better elucidate the direct regulatory roles of miRNAs on the stemness, growth, and differentiation of ovarian cancer-initiating cells. Other critical components of the tumor initiation and progression process, including enhanced proliferative capabilities, evading immunocompetent cells, uncontrolled cell division, and maintaining survival (63), and how miRNAs regulate each step will be discussed in more detail later.
MicroRNAs Regulate Proliferation, Cell Cycle, and Survivability of Ovarian Carcinomas
The ability of cancer cells to maintain active proliferation and survive in harsh and continuously changing environments are two of the most critical hallmarks of neoplastic diseases (64). Cell cycle progression is a multifaced process heavily guarded by several growth factors and suppressors, including cyclins and their protein kinases and inhibitors (65). These proteins regulate the cell quiescence (G0 phase), cell cycle re-entrance (G0-G1), and cell cycle progression through different phases (G1, S, G2, M) (Figure 2) Over the years, an increasing number of studies has aimed to understand the mechanisms of how miRNAs influence tumor suppressors and oncogenes to regulate cell proliferation and cell cycle progression pathways in ovarian cancers.
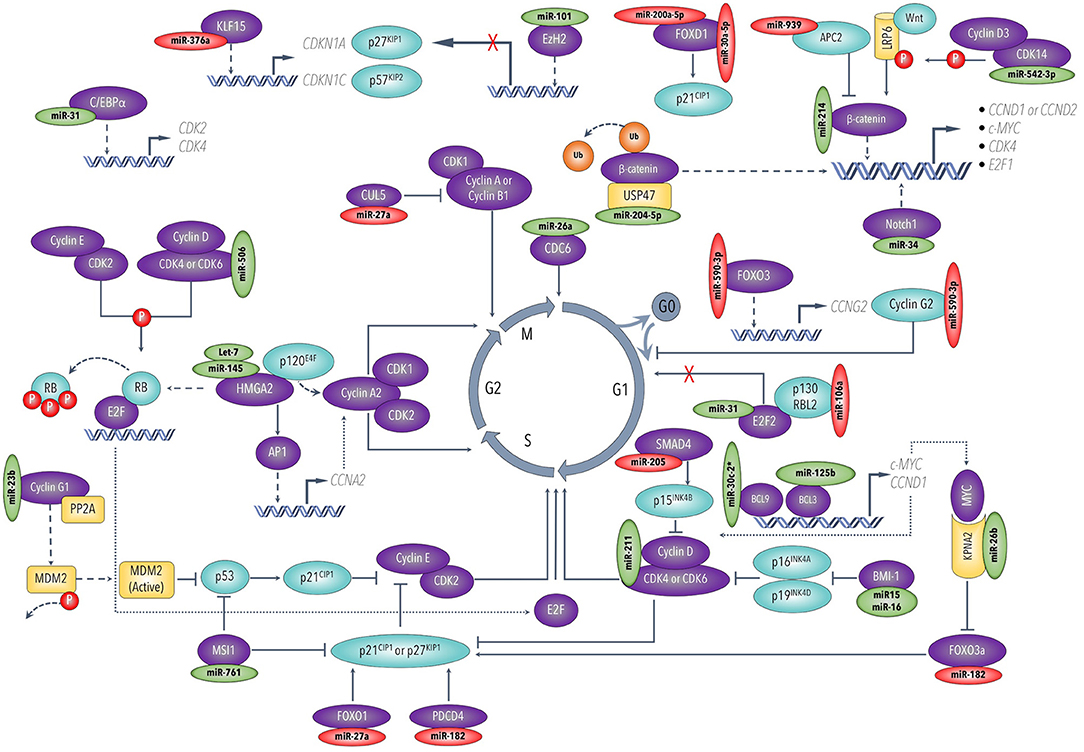
Figure 2. Cell cycle progression and major regulatory miRNAs in ovarian cancer. This figure was drawn based on the information from Liu et al. (183), Shi et al. (190), Qie et al. (221), and Liu et al. (224).
The cyclin-dependent kinases (CDKs), CDK4/6 and CDK14, have emerged as important targets for miRNAs that have decreased expression in ovarian cancer, namely, miR-506 (66), miR-211 (67), miR-542-3p (68). Ectopic expression of these tumor-suppressive miRNAs inhibits ovarian cancer cell proliferation and cell cycle progression through different phases by engaging multiple downstream pathways. Mechanistically, the abundant CDK4/6 can promote cell cycle progression via two major mechanisms, as illustrated in Figure 2. First, CDK4/6 phosphorylates and releases the retinoblastoma (Rb) tumor suppressor protein and thus enables E2F transcription factors to upregulate downstream targets involved in cell cycle transition from G1 into S phase. Second, CDK4/6 reactivates cyclin E-CDK2 kinase by sequestering CDK inhibitors p21CIP1 and p27KIP1 (69). In another study, miR15a/ miR-16 were reported to be downregulated in ovarian cell lines and primary ovarian tissues. The resultant overexpression of their target proto-oncogene Bmi-1 promoted cell cycle progression by suppressing CDK4/6 repressors p16INK4a and p19INK4d, and thus drive the cell cycle forward at G1/S phase (70). CDK14, on the other hand, is reported to activate several cell cycle-controlling targets, downstream of Wnt/β-catenin signaling pathway, by phosphorylating the co-receptor lipoprotein receptor-related protein 6 (LRP6) (71).
In addition to the increased expression of CDKs, miRNAs can also modulate the expression of several cell cycle-regulating cyclins directly or indirectly. For instance, Salem et al. recently reported that miR-590-3p enhanced ovarian cancer cell proliferation by directly binding and suppressing the cell cycle repressor, cyclin G2 (72). Furthermore, ovarian cancer cells perhaps “strategically” keep the tumor suppressor miR-23b at lower expression to maintain proliferative status by increasing the expression of its oncogenic target CCNG1 (cyclin G1) to sufficient levels to activate Mdm2 and ultimately override inhibitory effects of the wild-type p53 (73, 74). Other cyclins have been shown to be modulated indirectly when their upstream controlling proteins are directly affected by miRNAs. For example, several lines of evidence reported that overexpressed HMGA2 conversely correlates with the expression levels of its regulators let-7 and miR-145, in the ovarian cancer tissues and cell lines (53, 57, 75). As shown in Figure 2, HMGA2 can drive the cancer cell cycle forward by multiple mechanisms (56). Briefly, HMGA2 directly binds and suppresses p120E4F transcriptional repressor, thus inducing cyclin A2 expression, a cell cycle regulator at the S and G2/M phases. HMGA2 can also induce cyclin A2 transcription by upregulating its upstream regulator activator protein-1 (AP1). In addition, HMGA2 facilitates the cell cycle G1-S phase transition by releasing E2F1 transcription factors from the suppressor protein Rb. More recently, miR-27a-mediated inhibition of the E3 ubiquitin ligase Cullin-5 (CUL5) might result in maintaining high levels of cyclin A and B1, which are required for G2-M phase transition and cell proliferation (76).
In addition to the direct impacts of miRNAs on the cell cycle checkpoint regulators, other signaling targets upstream of the cell cycle and proliferation have been shown to be manipulated by miRNAs in ovarian cancer. For example, the class O of forkhead box transcription factors (FOXO) and the extracellular signal transducers, SMADs, exhibit suppressive roles by inducing cell cycle arrest (65). Mechanistically, FOXO1 inhibits cell proliferation, apparently by inducing the expression of the CDK inhibitor p21Cip1 (77). A recent study revealed that oncogenic miR-27a could stimulate ovarian cancer cell proliferation by suppressing the expression of FOXO1 (78). The tumor suppressor FOXO3 can also induce cell cycle arrest by increasing the transcription of CDK inhibitors p21CIP1 and p27KIP1 (79, 80), and cyclin G2 (72). Multiple studies suggest that ovarian cancer cells maintain lower levels of FOXO3 presumably by upregulating several oncogenic miRNAs, including miR-182 (81) and miR-590-3p (72).
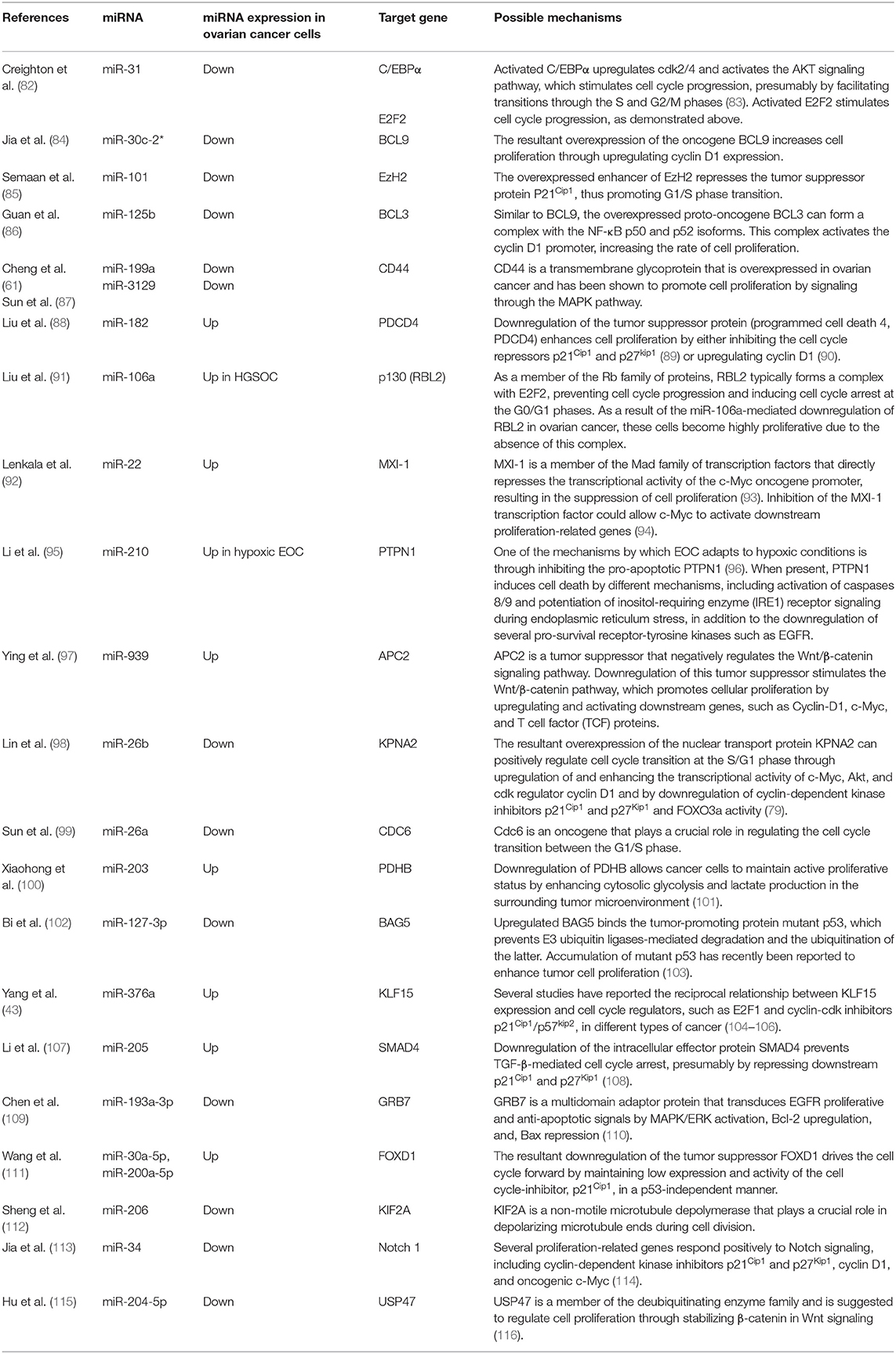
More studies that have investigated different miRNAs, their validated targets, and the downstream modulated targets that play essential roles in ovarian cancer proliferation and survival capabilities are listed in detail in Figures 2, 3. In addition, Table 2 summarizes more possible mechanisms by which miRNAs regulate the cell cycle progression and proliferation in ovarian cancer cells based on the reported targets. Since some of these mechanisms were not demonstrated in ovarian cancer cells per se, more studies are encouraged to validate that what has been shown in other cancers can be applied to ovarian cancer.
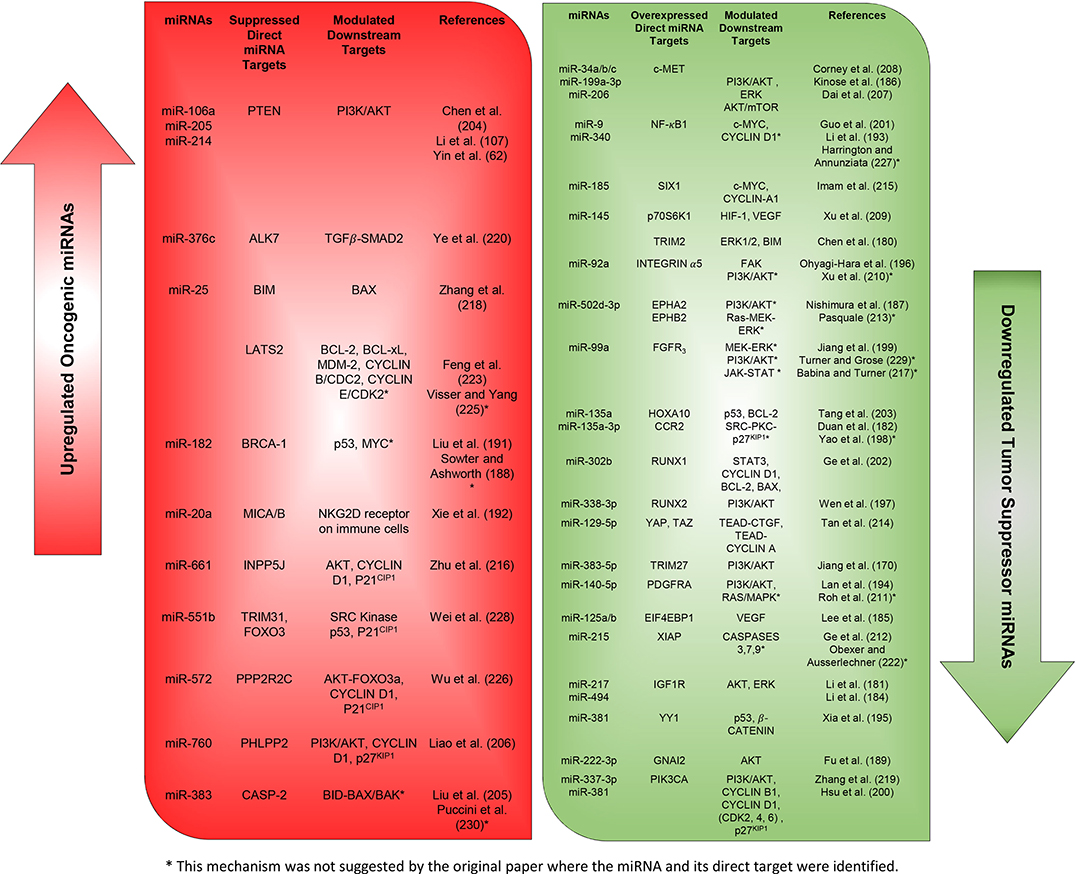
Figure 3. Expressions of oncogenic and tumor suppressor miRNAs and their reported downstream targets associated with cellular proliferation and survival in ovarian cancer.
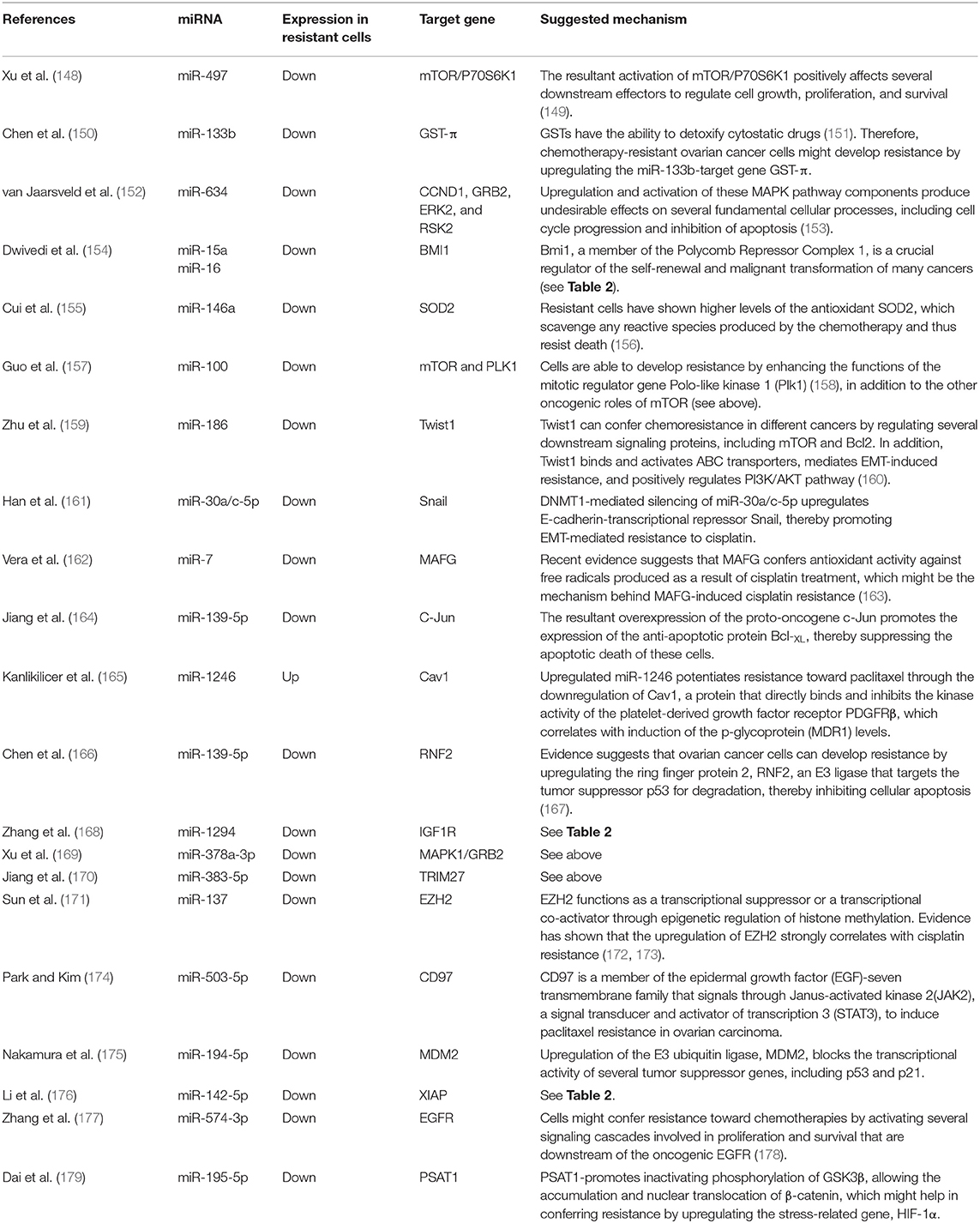
Roles of Micrornas in Ovarian Cancer Chemosensitivity and Resistance
miRNA-200 Family
The miR-200 family consists of five miRNA members (miR-141, miR-200a, miR-200b, miR-200c, and miR-429), whose expressions are reported to be significantly deregulated in different ovarian cancer tissues and cell lines (14, 19, 32, 35). Recent studies have linked miR-200 family members with the responsiveness of ovarian cancer patients to chemotherapeutic drugs. For example, p38α MAPK was previously shown to play an important role in reducing tumorigenesis by regulating cell proliferation, survival, and stress response (117, 118). Mateescu et al. identified p38α MAPK as a direct target for two of the most highly expressed miRNAs in ovarian cancer tissues, miR-141 and miR-200a (119). Both miR-141 and miR-200a increased paclitaxel sensitivity in a ROS-dependent manner by repressing p38α MAPK in vivo. More recently, a study by Liu et al. also reported that ectopic expression of miR-200a sensitized the ovarian cancer cell line OVCAR-3 to paclitaxel (120). MiR-200c is one of the most comprehensively investigated miRNAs in ovarian cancer. Although the majority of studies have shown that miR-200c is highly expressed in the tumor tissues as well as serum of ovarian cancer patients (32, 35), other studies have demonstrated that miR-200c was significantly downregulated in many advanced, poorly differentiated ovarian adenocarcinomas (121, 122). It seems that what controls the expression of these miRNAs are the disease stage, the type of specimen, and whether or not these cells have been exposed to treatment. Cochrane et al. reported that exogenous overexpression of miR-200c in different ovarian cancer cell lines resulted in a dramatic reduction in the expression of class III β-tubulin (TUBB3), a highly prevalent mechanism of resistance to microtubule-binding chemotherapies in many solid tumors (123–125). A study by Leskela et al. confirmed the previous findings by demonstrating that highly resistant ovarian tumors had high levels of the TUBB3 protein and significant downregulation in the expression of miR-141, miR-429 and miR-200c (126). Studies at the other end of the spectrum revealed that high expression of miR-141 and miR-200c was strongly correlated with ovarian cancer resistance to platinum-based therapy. Ectopic overexpression of miR-141 induced cisplatin resistance in ovarian cancer cells by directly suppressing the expression of the tumor suppressor Kelch-like ECH-associated protein 1 (KEAP1) and subsequently activating its downstream NF-κB pathway (127).
Epithelial-mesenchymal transition (EMT) is another biological process by which cancer cells confer resistance to chemotherapy (128). Increasing evidence suggests that the expression of key EMT-associated genes is directly regulated by several members of the miR-200 family. For instance, Zinc-finger E-box binding homeobox 1, ZEB1 (also known as δEF1), and zinc finger E-box-binding homeobox 2, ZEB2 (also known as SIP1), the upregulation of which facilitate EMT and tumor metastasis by downregulating E-cadherin, were identified as direct targets of all five members of the miR-200 family. Intriguingly, inhibition of these microRNAs was sufficient to induce EMT and reduce adhesion by upregulating E-cadherin transcriptional repressors, ZEB1, and/or SIP1 (121, 129). In agreement with these studies, two independent research groups have demonstrated that resorting to the expression of miR-429 and the other miR-200 family members induced mesenchymal-to-epithelial transition (MET), reverted the EMT phenotype, and sensitized different ovarian cancer cells to Taxol and platinum-based chemotherapies (130, 131). Furthermore, a study by Chen et al. demonstrated that miR-200c was significantly downregulated in the more aggressive and chemoresistant phenotype CD117+/CD44+ ovarian cancer stem cells compared with the CD117−/CD44− cells. Ectopic overexpression of miR-200c in the CD117+/CD44+ cells inhibited EMT and induced tumor suppression by directly repressing ZEB1 and Vimentin, which resulted in a subsequent upregulation of the E-cadherin expression (122). Collectively, these observations support the notion that the miR-200 family of microRNAs influences the sensitivity of ovarian cancer cells to conventional chemotherapies through multiple mechanisms, including, but not limited to, interference with cellular microtubule assembly, regulating NF-κB signaling, and the EMT/MET processes.
miRNA-214/199a Cluster
The upregulation of miR-214 in various human malignancies, including ovarian cancers, and its roles in chemosensitivity and resistance have been extensively studied and documented. One of the earliest works by Yang et al. demonstrated a significant upregulation of miR-214 expression (8.63-fold) in late-stage, high-grade tumors compared to the normal ovarian tissues. Mechanistic experiments revealed that miR-214 negatively regulated PTEN by binding to its 3′-UTR, leading to the repression of its translation and subsequent activation of its downstream Akt signaling cascade. The resulting activation of the AKT pathway by miR-214 promoted ovarian cancer cell survival and cisplatin resistance. Furthermore, miR-214 knockdown abrogated cisplatin resistance in the cisplatin-resistant cells (132). Since then, several studies have reported a reciprocal relationship between miR-214 expression and the degree of resistance to multiple therapies in numerous malignancies, including pancreatic (133) and gastric cancers (134, 135). Wang et al. have recently demonstrated that exosome-containing anti-miR-214 resensitized cisplatin-resistant gastric cancer cells in vitro and in vivo, serving as a promising treatment approach for refractory gastric cancer (136).
MiR-199a* (also known as miR-199a-5p) is another downregulated miRNA in ovarian cancer tissues relative to normal ovarian counterparts and has been implicated as playing an important role in ovarian cancer sensitivity to chemotherapeutics. In 2008, a study conducted by Chen et al. identified for the first time the functional role of miR-199a* as a direct inhibitor of IKKβ expression, a kinase that phosphorylates IκB proteins, causing their proteasomal degradation and subsequent nuclear translocation of NF-κB. They reported that Type I/MyD88 positive EOC cells had the capacity to develop resistance to cytotoxic drugs such as paclitaxel and TNF-α due to lower miR-199a* expression and higher IKKβ expression (137). In another study, HIF-1α and HIF-2α were identified as direct targets for miR-199a*. High expression of these two hypoxia-responsive factors and low expression of miR-199a* mediated ovarian cancer cell resistance to carboplatin. This observation was validated when exogenous overexpression of miR-199a* in the EOC cell line (A2780) disrupted cells' peritoneal seeding and sensitized them to carboplatin treatment in vivo (138). CD44+/CD117+ cells are ovarian cancer-initiating cells (CIC) that are characterized by their highly proliferative capabilities and their resistance to chemotherapies (58, 59). Cheng et al. found that miR-199a significantly enhanced the chemosensitivity of ovarian CICs to Adriamycin, cisplatin, and paclitaxel, and reduced the mRNA expression of the multidrug resistance gene ABCG2 (61). Most recently, Liu et al. have reported that miR-199b* (also known as miR-199b-5p) was significantly downregulated in cisplatin-resistant ovarian cancer cells relative to cisplatin-sensitive cells. They found that the loss of miR-199b-5p activated the JAG1-mediated Notch1 signaling cascade, a reported chemoresistance and survival-regulating pathway in different types of human malignancies (139, 140). Restoration of miR-199b-5p expression resensitized the ovarian cancer cells to cisplatin-induced cytotoxicity both in vitro and in vivo. Findings from these studies further insinuate the diversity of targets of these miRNAs and their pivotal roles in modulating cellular response to chemotherapies.
Let-7 Family
The let-7 (lethal-7) family of miRNAs consists of 13 human homologs that are located on nine different chromosomes (48, 141). Several studies have reported that the expression profiles of the let-7 family members were significantly reduced in various human cancer tissues and cell lines, including ovarian cancer (49–53, 57, 142). In addition to the roles of let-7 family members in the initiation and progression of ovarian cancer as mentioned above, studies have also shown a strong correlation between dysregulation in let-7 expression and the development of resistance to chemotherapeutic drugs. For example, Yang et al. have demonstrated that let-7i expression was significantly reduced in the tumor tissue samples derived from chemotherapy-resistant ovarian cancer patients. Loss-of-function (let-7i inhibition) and gain-of-function (let-7i overexpression) studies revealed that ovarian cancer resistance to cisplatin was significantly increased in the absence of let-7i, whereas ectopic expression of let-7i resensitized cells to treatment (142). Similarly, Boyerinas et al. reported that the sensitivity of multidrug resistant ovarian cancer cells to microtubule-targeting drugs (Taxanes) was increased by overexpressing let-7g in those cells. This effect was mediated by repressing IMP-1, which in turn causes destabilization of multidrug resistance (MDR1) at the mRNA and protein level, thereby increasing the sensitivity of the multidrug-resistant ovarian cancer cell to Taxanes but not carboplatin, a non-MDR1 substrate (143). Let-7e is another member of the let-7 family that has been reported recently to play an important role in the development of cisplatin resistance in ovarian cancer cells. Findings by Cai et al. demonstrated that let-7e expression was significantly downregulated in cisplatin-resistant human EOC cells compared to parental ones. Exogenous overexpression of let-7e resensitized the former to cisplatin by reducing the expression of cisplatin-resistant related proteins EZH2 and cyclin D1 (144). Deregulation of some members of the let-7 family of miRNAs have also been suggested to serve as potential biomarkers to predict the responsiveness of ovarian cancer patients to different chemotherapeutic agents. For instance, Lu et al. reported that ovarian cancer patients with higher let-7a expression did not respond well to paclitaxel/platinum combination therapy and had worse overall survival rates compared to patients with low let-7a expression (145). More recently, analysis of human EOC tissue specimens revealed that the tumor suppressors let-7g and let-7d-5p were strongly associated with acquired resistance to cisplatin (146, 147). Other individual miRNAs, their identified targets, and examples of the mechanisms by which these miRNAs could modulate the responsiveness of ovarian cancer cells to chemotherapy are listed in Table 3. Approximately 53% of the miRNAs reported in this review modulate ovarian cancer responsiveness to cisplatin, whereas ~25% play a significant role in cell resistance against taxanes-based therapy. As shown in Figure 4, members of the let-7 family, miR-214/199a cluster, and miR-200 family were among the 12 miRNAs that appear to be critical regulators of chemoresistance to cisplatin and paclitaxel. Furthermore, five chemoresistance-associated miRNAs in ovarian cancer patients (miR-141, miR-146a, miR-200c, miR-429, and miR-7) have been suggested to serve as novel prognostic biomarkers. Interestingly, only two miRNAs (miR-106a and miR-16) showed the ability to modulate proliferation and chemosensitivity of ovarian cancer cells and to function as prognosis indicators in ovarian cancer patients (Figure 5).
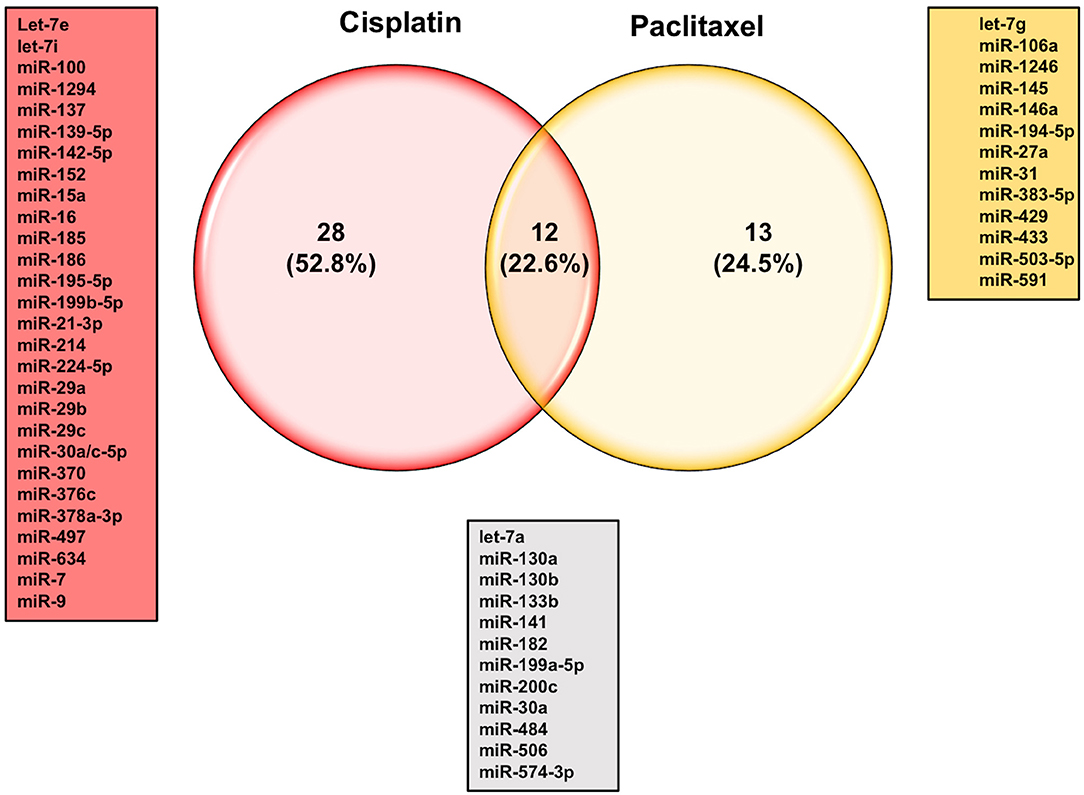
Figure 4. Venn diagram of reported miRNAs that strongly modulate ovarian cancer sensitivity to chemotherapeutic agents.
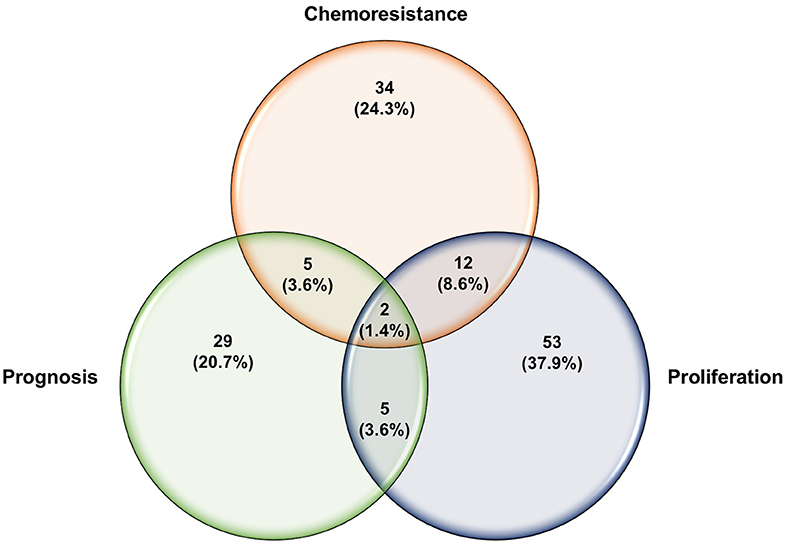
Figure 5. Venn diagram of overlapping miRNAs reported to regulate different aspects of ovarian carcinoma.
Concluding Remarks
The treatment of patients with ovarian cancer faces multiple challenges ranging from late diagnosis, due to the lack of reliable markers, to the development of resistance to current therapeutics owing to the heterogeneity of the disease. Over the last 20 years, substantial progress has been made in our understanding of how miRNAs regulate different cancer hallmarks and how we can utilize such knowledge to improve the diagnosis and prognosis of cancer patients. However, several challenges could affect the confidence in utilizing miRNAs as diagnostic and prognostic biomarkers, including diversified sampling methods, detection techniques, gender, and ethnicity or genetic background. More studies are needed with a larger number of patients to investigate the impact of each of the above factors on the identified miRNA signatures. Research on how miRNAs regulate the pathology of ovarian cancer is still in its infancy, representing <4% of the total published reports. The bulk of these studies deal with expression profiling, identification of target genes, determination of affected cellular pathways, resistance to conventional chemotherapeutics, and their utilization as therapeutic tools. As demonstrated in this review, overexpressing tumor suppressor miRNAs, using synthetic miRNAs mimics, or inhibiting the activity of oncogenic miRNAs, using anti-miRNA oligonucleotides, have a strong potential to serve as a therapeutic approach for the treatment of ovarian cancer.
Author Contributions
AA herby confirm sole responsibility for all information submitted in this manuscript and also declare my 100% contribution in this manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author is very grateful to the Deanship of Scientific Research and Research Center, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia.
Abbreviations
ALK7, activin receptor-like kinase-7; APC2, adenomatosis polyposis coli 2; ARID1A, AT-rich interaction domain 1A; BAG5, Bcl-2-associated athanogene 5; Bax, Bcl-2-associated X protein; BCL3, B-cell lymphoma 3; BCL9, B-cell CLL/lymphoma 9; Bim, Bcl-2 homologous 3; Bmi-1, polycomb ring finger; C/EBPα, CCAAT enhancer binding protein alpha; Cav1, caveolin-1; CCND1, cyclin D1; CCR2, C-C motif chemokine receptor 2; CDC6, cell division cycle 6; CTGF, connective tissue growth factor; CUL5, cullin5; E2F2, E2F transcription factor 2; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; EphA2, EPH receptor A2; EphB2, EPH receptor B2; EzH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; FAK, Focal adhesion kinase; FGFR3, fibroblast growth factor receptor 3; FOXD1, forkhead box D1; FOXO1, forkhead box O1; FOXO3, forkhead box O3; GNAI2, G protein alpha inhibiting activity polypeptide 2; GRB7, growth factor receptor bound protein 7; GSK3β, glycogen synthase kinase 3 beta; GST-π, glutathione S-transferase pi 1; HIF-1α, hypoxia-inducible factor 1 alpha; HMGA2, high mobility group AT-hook 2; HOXA10, homeobox A10; IGF1R, insulin like growth factor 1 receptor; INPP5J, inositol polyphosphate-5-phosphatase J; KIF2A, kinesin family member 2A; KLF15, Kruppel like factor 15; KPNA2, karyopherin subunit alpha 2; LATS2, large tumor suppressor kinase 2; LRP6, LDL receptor-related protein 6; MAFG, MAF bZIP transcription factor G; MAPK1, mitogen-activated protein kinase 1; MICA/B, MHC class I polypeptide-related sequences A/B; MSI1, Musashi RNA-binding protein1; mTOR, mechanistic target of rapamycin kinase; MXI-1, MAX-interacting protein 1; NF-κB1, nuclear factor kappa B subunit 1; NKG2D, killer cell lectin like receptor K1; OLFM4, olfactomedin 4; p130 (RBL2), retinoblastoma transcriptional corepressor like 2; p70S6K1, 70kDa ribosomal S6 kinases; PAK3, p21 (RAC1) activated kinase 3; PDCD4, programmed cell death 4; PDGFRA, platelet-derived growth factor receptor alpha; PDHB, pyruvate dehydrogenase E1 subunit beta; PHLPP2, PH domain and leucine-rich repeat protein phosphatase 2; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PPP2R2C, protein phosphatase 2 regulatory subunit B gamma; PSAT1, phosphoserine aminotransferase 1; PTEN, phosphatase and tensin homolog; PTPN1, protein tyrosine phosphatase non-receptor type 1; RSK2, ribosomal protein S6 kinase A3; RUNX1/2, runt-related transcription factor 1/2; SEMA4D, semaphorin 4D; Six1, SIX homeobox 1; SOD2, superoxide dismutase 2; STAT3, signal transducers and activators of transcription3; TAZ, tafazzin; TEAD, TEA domain; TGF-β, transforming growth factor β; TP53, tumor protein p53; TRIM2, tripartite motif containing 2; USP47, ubiquitin specific peptidase 47; VEGF, vascular endothelial growth factor; XIAP, X-linked inhibitor of apoptosis; YAP, Yes-associated Protein.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
3. Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA. (2011) 305:2295–303. doi: 10.1001/jama.2011.766
4. Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol. (2009) 4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246
5. Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. (2009) 9:415–28. doi: 10.1038/nrc2644
6. Ledermann JA, Raja FA, Fotopoulou C, Gonzalez-Martin A, Colombo N, Sessa C, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2013) 24(Suppl 6):vi24–32. doi: 10.1093/annonc/mdt333
7. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucl Acids Res. (2019) 47:D155–62. doi: 10.1093/nar/gky1141
8. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. (2004) 5:522–31. doi: 10.1038/nrg1379
9. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
10. Friedlander MR, Lizano E, Houben AJ, Bezdan D, Banez-Coronel M, Kudla G, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. (2014) 15:R57. doi: 10.1186/gb-2014-15-4-r57
11. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. (2009) 10:704–14. doi: 10.1038/nrg2634
12. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. (2014) 20:460–9. doi: 10.1016/j.molmed.2014.06.005
13. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. (2002) 99:15524–9. doi: 10.1073/pnas.242606799
14. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. (2007) 67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936
15. Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. (2008) 105:7004–9. doi: 10.1073/pnas.0801615105
16. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. (2005) 120:635–47. doi: 10.1016/j.cell.2005.01.014
17. Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. (2006) 9:435–43. doi: 10.1016/j.ccr.2006.04.020
18. Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. (2008) 7:35. doi: 10.1186/1476-4598-7-35
19. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. (2008) 14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731
20. Calura E, Fruscio R, Paracchini L, Bignotti E, Ravaggi A, Martini P, et al. MiRNA landscape in stage I epithelial ovarian cancer defines the histotype specificities. Clin Cancer Res. (2013) 19:4114–23. doi: 10.1158/1078-0432.CCR-13-0360
21. Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. (2014) 14:80. doi: 10.1186/1471-2407-14-80
22. Bovell LC, Shanmugam C, Putcha BD, Katkoori VR, Zhang B, Bae S, et al. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. (2013) 19:3955–65. doi: 10.1158/1078-0432.CCR-12-3302
23. Nassar FJ, Talhouk R, Zgheib NK, Tfayli A, El Sabban M, El Saghir NS, et al. microRNA expression in ethnic specific early stage breast cancer: an integration and comparative analysis. Sci Rep. (2017) 7:16829. doi: 10.1038/s41598-017-16978-y
24. Li E, Ji P, Ouyang N, Zhang Y, Wang XY, Rubin DC, et al. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol. (2014) 45:587–94. doi: 10.3892/ijo.2014.2469
25. Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. (2014) 18:371–90. doi: 10.1111/jcmm.12236
26. Sanz-Rubio D, Martin-Burriel I, Gil A, Cubero P, Forner M, Khalyfa A, et al. Stability of circulating exosomal miRNAs in healthy subjects. Sci Rep. (2018) 8:10306. doi: 10.1038/s41598-018-28748-5
27. Ando H, Hirose M, Kurosawa G, Impey S, Mikoshiba K. Time-lapse imaging of microRNA activity reveals the kinetics of microRNA activation in single living cells. Sci Rep. (2017) 7:12642. doi: 10.1038/s41598-017-12879-2
28. Reichholf B, Herzog VA, Fasching N, Manzenreither RA, Sowemimo I, Ameres SL. Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol Cell. (2019) 75:756–68 e757. doi: 10.1016/j.molcel.2019.06.018
29. Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. (2018) 10:59. doi: 10.1186/s13148-018-0492-1
30. Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. (2018) 9:4319. doi: 10.1038/s41467-018-06434-4
31. Zhao L, Wang W, Xu L, Yi T, Zhao X, Wei Y, et al. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene. (2019) 38:2305–19. doi: 10.1038/s41388-018-0577-5
32. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. (2008) 110:13–21. doi: 10.1016/j.ygyno.2008.04.033
33. Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. (2009) 112:55–9. doi: 10.1016/j.ygyno.2008.08.036
34. Hausler SF, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, et al. Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. (2010) 103:693–700. doi: 10.1038/sj.bjc.6605833
35. Kan CW, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. (2012) 12:627. doi: 10.1186/1471-2407-12-627
36. Suryawanshi S, Vlad AM, Lin HM, Mantia-Smaldone G, Laskey R, Lee M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. (2013) 19:1213–24. doi: 10.1158/1078-0432.CCR-12-2726
37. Shapira I, Oswald M, Lovecchio J, Khalili H, Menzin A, Whyte J, et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer. (2014) 110:976–83. doi: 10.1038/bjc.2013.795
38. Gao YC, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. (2015) 36:4843–50. doi: 10.1007/s13277-015-3138-3
39. Liang H, Jiang Z, Xie G, Lu Y. Serum microRNA-145 as a novel biomarker in human ovarian cancer. Tumour Biol. (2015) 36:5305–13. doi: 10.1007/s13277-015-3191-y
40. Langhe R, Norris L, Saadeh FA, Blackshields G, Varley R, Harrison A, et al. A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. (2015) 356(2 Pt B):628–36. doi: 10.1016/j.canlet.2014.10.010
41. Meng X, Joosse SA, Muller V, Trillsch F, Milde-Langosch K, Mahner S, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. (2015) 113:1358–66. doi: 10.1038/bjc.2015.340
42. Meng X, Muller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. (2016) 7:16923–35. doi: 10.18632/oncotarget.7850
43. Yang L, Wei QM, Zhang XW, Sheng Q, Yan XT. MiR-376a promotion of proliferation and metastases in ovarian cancer: potential role as a biomarker. Life Sci. (2017) 173:62–7. doi: 10.1016/j.lfs.2016.12.007
44. Todeschini P, Salviato E, Paracchini L, Ferracin M, Petrillo M, Zanotti L, et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: a validation across two independent cohorts. Cancer Lett. (2017) 388:320–7. doi: 10.1016/j.canlet.2016.12.017
45. Yokoi A, Yoshioka Y, Hirakawa A, Yamamoto Y, Ishikawa M, Ikeda SI, et al. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget. (2017) 8:89811–23. doi: 10.18632/oncotarget.20688
46. Shah JS, Gard GB, Yang J, Maidens J, Valmadre S, Soon PS, et al. Combining serum microRNA and CA-125 as prognostic indicators of preoperative surgical outcome in women with high-grade serous ovarian cancer. Gynecol Oncol. (2018) 148:181–8. doi: 10.1016/j.ygyno.2017.11.005
47. Kim S, Choi MC, Jeong JY, Hwang S, Jung SG, Joo WD, et al. Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J Cancer. (2019) 10:1958–67. doi: 10.7150/jca.30231
48. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. (2008) 18:505–16. doi: 10.1016/j.tcb.2008.07.007
49. Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. (2010) 17:F19–36. doi: 10.1677/ERC-09-0184
50. Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. (2008) 68:2587–91. doi: 10.1158/0008-5472.CAN-08-0264
51. Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. (2007) 21:1025–30. doi: 10.1101/gad.1540407
52. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. (2007) 315:1576–9. doi: 10.1126/science.1137999
53. Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. (2007) 6:2585–90. doi: 10.4161/cc.6.21.4845
54. Mahajan A, Liu Z, Gellert L, Zou X, Yang G, Lee P, et al. HMGA2: a biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Mod Pathol. (2010) 23:673–81. doi: 10.1038/modpathol.2010.49
55. Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, et al. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. (2011) 71:349–59. doi: 10.1158/0008-5472.CAN-10-2550
56. Zhang S, Mo Q, Wang X. Oncological role of HMGA2 (review). Int J Oncol. (2019) 55:775–88. doi: 10.3892/ijo.2019.4856
57. Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. (2007) 104:11400–5. doi: 10.1073/pnas.0704372104
58. Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. (2008) 68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364
59. Liu T, Cheng W, Lai D, Huang Y, Guo L. Characterization of primary ovarian cancer cells in different culture systems. Oncol Rep. (2010) 23:1277–84. doi: 10.3892/or_00000761
60. Ma L, Lai D, Liu T, Cheng W, Guo L. Cancer stem-like cells can be isolated with drug selection in human ovarian cancer cell line SKOV3. Acta Biochim Biophys Sin. (2010) 42:593–602. doi: 10.1093/abbs/gmq067
61. Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. (2012) 279:2047–59. doi: 10.1111/j.1742-4658.2012.08589.x
62. Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. (2010) 29:3545–53. doi: 10.1038/onc.2010.111
63. Tysnes BB, Bjerkvig R. Cancer initiation and progression: involvement of stem cells and the microenvironment. Biochim Biophys Acta. (2007) 1775:283–97. doi: 10.1016/j.bbcan.2007.01.001
64. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
65. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. (2017) 17:93–115. doi: 10.1038/nrc.2016.138
66. Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, et al. MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol. (2014) 233:308–18. doi: 10.1002/path.4348
67. Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. (2015) 14:57. doi: 10.1186/s12943-015-0322-4
68. Li J, Shao W, Feng H. MiR-542-3p, a microRNA targeting CDK14, suppresses cell proliferation, invasiveness, and tumorigenesis of epithelial ovarian cancer. Biomed Pharmacother. (2019) 110:850–6. doi: 10.1016/j.biopha.2018.11.104
69. Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. (2004) 18:2699–711. doi: 10.1101/gad.1256504
70. Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. (2009) 69:9090–5. doi: 10.1158/0008-5472.CAN-09-2552
71. Ji Q, Xu X, Li L, Goodman SB, Bi W, Xu M, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. (2017) 8:e3103. doi: 10.1038/cddis.2017.499
72. Salem M, Shan Y, Bernaudo S, Peng C. miR-590-3p targets cyclin G2 and FOXO3 to promote ovarian cancer cell proliferation, invasion, and spheroid formation. Int J Mol Sci. (2019) 20:1810. doi: 10.3390/ijms20081810
73. Yan J, Jiang JY, Meng XN, Xiu YL, Zong ZH. MiR-23b targets cyclin G1 and suppresses ovarian cancer tumorigenesis and progression. J Exp Clin Cancer Res. (2016) 35:31. doi: 10.1186/s13046-016-0307-1
74. Al-Shihabi A, Chawla SP, Hall FL, Gordon EM. Exploiting oncogenic drivers along the CCNG1 pathway for cancer therapy and gene therapy. Mol Ther Oncolyt. (2018) 11:122–6. doi: 10.1016/j.omto.2018.11.002
75. Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH, et al. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. (2015) 356:937–45. doi: 10.1016/j.canlet.2014.11.011
76. Si L, Jia Y, Lin R, Jian W, Yu Q, Yang S. MicroRNA-27a regulates the proliferation, chemosensitivity and invasion of human ovarian cancer cell lines by targeting Cullin 5. Arch Biochem Biophys. (2019) 668:9–15. doi: 10.1016/j.abb.2019.04.009
77. Prasad SB, Yadav SS, Das M, Govardhan HB, Pandey LK, Singh S, et al. Down regulation of FOXO1 promotes cell proliferation in cervical cancer. J Cancer. (2014) 5:655–62. doi: 10.7150/jca.6554
78. Wang Z, Ji G, Wu Q, Feng S, Zhao Y, Cao Z, et al. Integrated microarray meta-analysis identifies miRNA-27a as an oncogene in ovarian cancer by inhibiting FOXO1. Life Sci. (2018) 210:263–70. doi: 10.1016/j.lfs.2018.08.043
79. Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, et al. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis. (2013) 4:e745. doi: 10.1038/cddis.2013.256
80. Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, et al. Critical role of FOXO3a in carcinogenesis. Mol Cancer. (2018) 17:104. doi: 10.1186/s12943-018-0856-3
81. Levanon K, Sapoznik S, Bahar-Shany K, Brand H, Shapira-Frommer R, Korach J, et al. FOXO3a loss is a frequent early event in high-grade pelvic serous carcinogenesis. Oncogene. (2014) 33:4424–32. doi: 10.1038/onc.2013.394
82. Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. (2010) 70:1906–15. doi: 10.1158/0008-5472.CAN-09-3875
83. Yin H, Lowery M, Glass J. In prostate cancer C/EBPalpha promotes cell growth by the loss of interactions with CDK2, CDK4, and E2F and by activation of AKT. Prostate. (2009) 69:1001–16. doi: 10.1002/pros.20947
84. Jia W, Eneh JO, Ratnaparkhe S, Altman MK, Murph MM. MicroRNA-30c-2* expressed in ovarian cancer cells suppresses growth factor-induced cellular proliferation and downregulates the oncogene BCL9. Mol Cancer Res. (2011) 9:1732–45. doi: 10.1158/1541-7786.MCR-11-0245
85. Semaan A, Qazi AM, Seward S, Chamala S, Bryant CS, Kumar S, et al. MicroRNA-101 inhibits growth of epithelial ovarian cancer by relieving chromatin-mediated transcriptional repression of p21[waf(1)/cip(1)]. Pharm Res. (2011) 28:3079–90. doi: 10.1007/s11095-011-0547-x
86. Guan Y, Yao H, Zheng Z, Qiu G, Sun K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. (2011) 128:2274–83. doi: 10.1002/ijc.25575
87. Sun X, Cui M, Tong L, Zhang A, Wang K. Upregulation of microRNA-3129 suppresses epithelial ovarian cancer through CD44. Cancer Gene Ther. (2018) 25:317–25. doi: 10.1038/s41417-018-0026-1
88. Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem. (2013) 114:1464–73. doi: 10.1002/jcb.24488
89. Wei N, Liu SS, Chan KK, Ngan HY. Tumour suppressive function and modulation of programmed cell death 4 (PDCD4) in ovarian cancer. PLoS ONE. (2012) 7:e30311. doi: 10.1371/journal.pone.0030311
90. Guo X, Li W, Wang Q, Yang HS. AKT activation by Pdcd4 knockdown up-regulates cyclin D1 expression and promotes cell proliferation. Genes Cancer. (2011) 2:818–28. doi: 10.1177/1947601911431082
91. Liu Z, Gersbach E, Zhang X, Xu X, Dong R, Lee P, et al. miR-106a represses the Rb tumor suppressor p130 to regulate cellular proliferation and differentiation in high-grade serous ovarian carcinoma. Mol Cancer Res. (2013) 11:1314–25. doi: 10.1158/1541-7786.MCR-13-0131
92. Lenkala D, LaCroix B, Gamazon ER, Geeleher P, Im HK, Huang RS. The impact of microRNA expression on cellular proliferation. Hum Genet. (2014) 133:931–8. doi: 10.1007/s00439-014-1434-4
93. Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. (1993) 72:223–32. doi: 10.1016/0092-8674(93)90662-A
94. Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. (2018) 3:5. doi: 10.1038/s41392-018-0008-7
95. Li L, Huang K, You Y, Fu X, Hu L, Song L, et al. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int J Oncol. (2014) 44:2111–20. doi: 10.3892/ijo.2014.2368
96. Banh RS, Iorio C, Marcotte R, Xu Y, Cojocari D, Rahman AA, et al. PTP1B controls non-mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nat Cell Biol. (2016) 18:803–13. doi: 10.1038/ncb3376
97. Ying X, Li-ya Q, Feng Z, Yin W, Ji-hong L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed Pharmacother. (2015) 71:64–9. doi: 10.1016/j.biopha.2015.02.020
98. Lin J, Zhang L, Huang H, Huang Y, Huang L, Wang J, et al. MiR-26b/KPNA2 axis inhibits epithelial ovarian carcinoma proliferation and metastasis through downregulating OCT4. Oncotarget. (2015) 6:23793–806. doi: 10.18632/oncotarget.4363
99. Sun TY, Xie HJ, He H, Li Z, Kong LF. miR-26a inhibits the proliferation of ovarian cancer cells via regulating CDC6 expression. Am J Transl Res. (2016) 8:1037–46.
100. Xiaohong Z, Lichun F, Na X, Kejian Z, Xiaolan X, Shaosheng W. MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumour Biol. (2016) 37:14989–97. doi: 10.1007/s13277-016-5415-1
101. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
102. Bi L, Yang Q, Yuan J, Miao Q, Duan L, Li F, et al. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep. (2016) 36:2563–70. doi: 10.3892/or.2016.5055
103. Yue X, Zhao Y, Huang G, Li J, Zhu J, Feng Z, et al. A novel mutant p53 binding partner BAG5 stabilizes mutant p53 and promotes mutant p53 GOFs in tumorigenesis. Cell Discov. (2016) 2:16039. doi: 10.1038/celldisc.2016.39
104. Ray S, Pollard JW. KLF15 negatively regulates estrogen-induced epithelial cell proliferation by inhibition of DNA replication licensing. Proc Natl Acad Sci USA. (2012) 109:E1334–43. doi: 10.1073/pnas.1118515109
105. Yoda T, McNamara KM, Miki Y, Onodera Y, Takagi K, Nakamura Y, et al. KLF15 in breast cancer: a novel tumor suppressor? Cell Oncol. (2015) 38:227–35. doi: 10.1007/s13402-015-0226-8
106. Sun C, Ma P, Wang Y, Liu W, Chen Q, Pan Y, et al. KLF15 inhibits cell proliferation in gastric cancer cells via up-regulating CDKN1A/p21 and CDKN1C/p57 expression. Dig Dis Sci. (2017) 62:1518–26. doi: 10.1007/s10620-017-4558-2
107. Li J, Hu K, Gong G, Zhu D, Wang Y, Liu H, et al. Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci Rep. (2017) 7:41330. doi: 10.1038/srep41330
108. Zhao M, Mishra L, Deng CX. The role of TGF-beta/SMAD4 signaling in cancer. Int J Biol Sci. (2018) 14:111–23. doi: 10.7150/ijbs.23230
109. Chen K, Liu MX, Mak CS, Yung MM, Leung TH, Xu D, et al. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics. (2018) 8:423–36. doi: 10.7150/thno.22377
110. Chu PY, Tai YL, Shen TL. Grb7, a critical mediator of EGFR/ErbB signaling, in Cancer Development and as a Potential Therapeutic Target. Cells (2019) 8:435. doi: 10.3390/cells8050435
111. Wang Y, Qiu C, Lu N, Liu Z, Jin C, Sun C, et al. FOXD1 is targeted by miR-30a-5p and miR-200a-5p and suppresses the proliferation of human ovarian carcinoma cells by promoting p21 expression in a p53-independent manner. Int J Oncol. (2018) 52:2130–42. doi: 10.3892/ijo.2018.4359
112. Sheng N, Xu YZ, Xi QH, Jiang HY, Wang CY, Zhang Y, et al. Overexpression of KIF2A is Suppressed by miR-206 and Associated with Poor Prognosis in Ovarian Cancer. Cell Physiol Biochem. (2018) 50:810–22. doi: 10.1159/000494467
113. Jia Y, Lin R, Jin H, Si L, Jian W, Yu Q, et al. MicroRNA-34 suppresses proliferation of human ovarian cancer cells by triggering autophagy and apoptosis and inhibits cell invasion by targeting Notch 1. Biochimie. (2019) 160:193–9. doi: 10.1016/j.biochi.2019.03.011
114. Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S, et al. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer Lett. (2015) 369:20–7. doi: 10.1016/j.canlet.2015.07.048
115. Hu L, Kolibaba H, Zhang S, Cao M, Niu H, Mei H, et al. MicroRNA-204-5p inhibits ovarian cancer cell proliferation by down-regulating USP47. Cell Transplant. (2019) 28:51S–8S. doi: 10.1177/0963689719877372
116. Shi J, Liu Y, Xu X, Zhang W, Yu T, Jia J, et al. Deubiquitinase USP47/UBP64E regulates beta-catenin ubiquitination and degradation and plays a positive role in Wnt signaling. Mol Cell Biol. (2015) 35:3301–11. doi: 10.1128/MCB.00373-15
117. Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. (2009) 9:537–49. doi: 10.1038/nrc2694
118. Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. (2010) 1802:396–405. doi: 10.1016/j.bbadis.2009.12.009
119. Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. (2011) 17:1627–35. doi: 10.1038/nm.2512
120. Liu N, Zhong L, Zeng J, Zhang X, Yang Q, Liao D, et al. Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer. Neoplasma. (2015) 62:550–9. doi: 10.4149/neo_2015_066
121. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. (2008) 10:593–601. doi: 10.1038/ncb1722
122. Chen D, Zhang Y, Wang J, Chen J, Yang C, Cai K, et al. MicroRNA-200c overexpression inhibits tumorigenicity and metastasis of CD117+CD44+ ovarian cancer stem cells by regulating epithelial-mesenchymal transition. J Ovarian Res. (2013) 6:50. doi: 10.1186/1757-2215-6-50
123. Tommasi S, Mangia A, Lacalamita R, Bellizzi A, Fedele V, Chiriatti A, et al. Cytoskeleton and paclitaxel sensitivity in breast cancer: the role of beta-tubulins. Int J Cancer. (2007) 120:2078–85. doi: 10.1002/ijc.22557
124. Seve P, Dumontet C. Is class III beta-tubulin a predictive factor in patients receiving tubulin-binding agents? Lancet Oncol. (2008) 9:168–75. doi: 10.1016/S1470-2045(08)70029-9
125. Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. (2009) 8:1055–66. doi: 10.1158/1535-7163.MCT-08-1046
126. Leskela S, Leandro-Garcia LJ, Mendiola M, Barriuso J, Inglada-Perez L, Munoz I, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. (2011) 18:85–95. doi: 10.1677/ERC-10-0148
127. van Jaarsveld MT, Helleman J, Boersma AW, van Kuijk PF, van Ijcken WF, Despierre E, et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. (2013) 32:4284–93. doi: 10.1038/onc.2012.433
128. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. (2017) 14:611–29. doi: 10.1038/nrclinonc.2017.44
129. Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. (2011) 121:200–5. doi: 10.1016/j.ygyno.2010.12.339
130. Wang L, Mezencev R, Svajdler M, Benigno BB, McDonald JF. Ectopic over-expression of miR-429 induces mesenchymal-to-epithelial transition (MET) and increased drug sensitivity in metastasizing ovarian cancer cells. Gynecol Oncol. (2014) 134:96–103. doi: 10.1016/j.ygyno.2014.04.055
131. Brozovic A, Duran GE, Wang YC, Francisco EB, Sikic BI. The miR-200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR-3 and MES-OV cells. Mol Oncol. (2015) 9:1678–93. doi: 10.1016/j.molonc.2015.04.015
132. Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. (2008) 68:425–33. doi: 10.1158/0008-5472.CAN-07-2488
133. Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. (2010) 3:46. doi: 10.1186/1756-8722-3-46
134. Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. (2013) 13:68. doi: 10.1186/1475-2867-13-68
135. Zhang KC, Xi HQ, Cui JX, Shen WS, Li JY, Wei B, et al. Hemolysis-free plasma miR-214 as novel biomarker of gastric cancer and is correlated with distant metastasis. Am J Cancer Res. (2015) 5:821–9.
136. Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, et al. Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol Ther. (2018) 26:774–83. doi: 10.1016/j.ymthe.2018.01.001
137. Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. (2008) 27:4712–23. doi: 10.1038/onc.2008.112
138. Joshi HP, Subramanian IV, Schnettler EK, Ghosh G, Rupaimoole R, Evans C, et al. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proc Natl Acad Sci USA. (2014) 111:5331–6. doi: 10.1073/pnas.1317242111
139. Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. (2013) 34:1420–30. doi: 10.1093/carcin/bgt127
140. Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. (2014) 5:944–58. doi: 10.18632/oncotarget.1458
141. Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. (2006) 127:1193–207. doi: 10.1016/j.cell.2006.10.040
142. Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. (2008) 68:10307–14. doi: 10.1158/0008-5472.CAN-08-1954
143. Boyerinas B, Park SM, Murmann AE, Gwin K, Montag AG, Zillhardt M, et al. Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of multidrug resistance 1. Int J Cancer. (2012) 130:1787–97. doi: 10.1002/ijc.26190
144. Cai J, Yang C, Yang Q, Ding H, Jia J, Guo J, et al. Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis. (2013) 2:e75. doi: 10.1038/oncsis.2013.39
145. Lu L, Schwartz P, Scarampi L, Rutherford T, Canuto EM, Yu H, et al. MicroRNA let-7a: a potential marker for selection of paclitaxel in ovarian cancer management. Gynecol Oncol. (2011) 122:366–71. doi: 10.1016/j.ygyno.2011.04.033
146. Biamonte F, Santamaria G, Sacco A, Perrone FM, Di Cello A, Battaglia AM, et al. MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer. Sci Rep. (2019) 9:5668. doi: 10.1038/s41598-019-42221-x
147. Chen YN, Ren CC, Yang L, Nai MM, Xu YM, Zhang F, et al. MicroRNA let7d5p rescues ovarian cancer cell apoptosis and restores chemosensitivity by regulating the p53 signaling pathway via HMGA1. Int J Oncol. (2019) 54:1771–84. doi: 10.3892/ijo.2019.4731
148. Xu S, Fu GB, Tao Z, OuYang J, Kong F, Jiang BH, et al. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. (2015) 6:26457–71. doi: 10.18632/oncotarget.4762
149. Fenton TR Gout IT. Functions and regulation of the 70kDa ribosomal S6 kinases. Int J Biochem Cell Biol. (2011) 43:47–59. doi: 10.1016/j.biocel.2010.09.018
150. Chen S, Jiao JW, Sun KX, Zong ZH, Zhao Y. MicroRNA-133b targets glutathione S-transferase pi expression to increase ovarian cancer cell sensitivity to chemotherapy drugs. Drug Des Devel Ther. (2015) 9:5225–35. doi: 10.2147/DDDT.S87526
151. Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. (2003) 22:7369–75. doi: 10.1038/sj.onc.1206940
152. van Jaarsveld MT, van Kuijk PF, Boersma AW, Helleman J, van IWF, Mathijssen RH, et al. miR-634 restores drug sensitivity in resistant ovarian cancer cells by targeting the Ras-MAPK pathway. Mol Cancer. (2015) 14:196. doi: 10.1186/s12943-015-0464-4
153. Achkar IW, Abdulrahman N, Al-Sulaiti H, Joseph JM, Uddin S, Mraiche F. Cisplatin based therapy: the role of the mitogen activated protein kinase signaling pathway. J Transl Med. (2018) 16:96. doi: 10.1186/s12967-018-1471-1
154. Dwivedi SK, Mustafi SB, Mangala LS, Jiang D, Pradeep S, Rodriguez-Aguayo C, et al. Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget. (2016) 7:15093–104. doi: 10.18632/oncotarget.7618
155. Cui Y, She K, Tian D, Zhang P, Xin X. miR-146a inhibits proliferation and enhances chemosensitivity in epithelial ovarian cancer via reduction of SOD2. Oncol Res. (2016) 23:275–82. doi: 10.3727/096504016X14562725373798
156. Kim EK, Jang M, Song MJ, Kim D, Kim Y, Jang HH. Redox-mediated mechanism of chemoresistance in cancer cells. Antioxidants. (2019) 8:471. doi: 10.3390/antiox8100471
157. Guo P, Xiong X, Zhang S, Peng D. miR-100 resensitizes resistant epithelial ovarian cancer to cisplatin. Oncol Rep. (2016) 36:3552–8. doi: 10.3892/or.2016.5140
158. Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. (2004) 5:429–40. doi: 10.1038/nrm1401
159. Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. (2016) 35:323–32. doi: 10.1038/onc.2015.84
160. Zhao Z, Rahman MA, Chen ZG, Shin DM. Multiple biological functions of Twist1 in various cancers. Oncotarget. (2017) 8:20380–93. doi: 10.18632/oncotarget.14608
161. Han X, Zhen S, Ye Z, Lu J, Wang L, Li P, et al. A feedback loop between miR-30a/c-5p and DNMT1 mediates cisplatin resistance in ovarian cancer cells. Cell Physiol Biochem. (2017) 41:973–86. doi: 10.1159/000460618
162. Vera O, Jimenez J, Pernia O, Rodriguez-Antolin C, Rodriguez C, Sanchez Cabo F, et al. DNA methylation of miR-7 is a mechanism involved in platinum response through MAFG overexpression in cancer cells. Theranostics. (2017) 7:4118–34. doi: 10.7150/thno.20112
163. Vera-Puente O, Rodriguez-Antolin C, Salgado-Figueroa A, Michalska P, Pernia O, Reid BM, et al. MAFG is a potential therapeutic target to restore chemosensitivity in cisplatin-resistant cancer cells by increasing reactive oxygen species. Transl Res. (2018) 200:1–17. doi: 10.1016/j.trsl.2018.06.005
164. Jiang Y, Jiang J, Jia H, Qiao Z, Zhang J. Recovery of miR-139-5p in ovarian cancer reverses cisplatin resistance by targeting C-jun. Cell Physiol Biochem. (2018) 51:129–41. doi: 10.1159/000495169
165. Kanlikilicer P, Bayraktar R, Denizli M, Rashed MH, Ivan C, Aslan B, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine. (2018) 38:100–12. doi: 10.1016/j.ebiom.2018.11.004
166. Chen Y, Cao XY, Li YN, Qiu YY, Li YN, Li W, et al. Reversal of cisplatin resistance by microRNA-139-5p-independent RNF2 downregulation and MAPK inhibition in ovarian cancer. Am J Physiol Cell Physiol. (2018) 315:C225–35. doi: 10.1152/ajpcell.00283.2017
167. Su WJ, Fang JS, Cheng F, Liu C, Zhou F, Zhang J. RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc Natl Acad Sci USA. (2013) 110:1720–5. doi: 10.1073/pnas.1211604110
168. Zhang Y, Huang S, Guo Y, Li L. MiR-1294 confers cisplatin resistance in ovarian cancer cells by targeting IGF1R. Biomed Pharmacother. (2018) 106:1357–63. doi: 10.1016/j.biopha.2018.07.059
169. Xu ZH, Yao TZ, Liu W. miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed Pharmacother. (2018) 107:1410–7. doi: 10.1016/j.biopha.2018.08.132
170. Jiang J, Xie C, Liu Y, Shi Q, Chen Y. Up-regulation of miR-383-5p suppresses proliferation and enhances chemosensitivity in ovarian cancer cells by targeting TRIM27. Biomed Pharmacother. (2019) 109:595–601. doi: 10.1016/j.biopha.2018.10.148
171. Sun J, Cai X, Yung MM, Zhou W, Li J, Zhang Y, et al. miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer. Oncogene. (2019) 38:564–80. doi: 10.1038/s41388-018-0459-x
172. Hu S, Yu L, Li Z, Shen Y, Wang J, Cai J, et al. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol Ther. (2010) 10:788–95. doi: 10.4161/cbt.10.8.12913
173. Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. (2018) 6:10. doi: 10.1186/s40364-018-0122-2
174. Park GB, Kim D. MicroRNA-503-5p inhibits the CD97-mediated JAK2/STAT3 pathway in metastatic or paclitaxel-resistant ovarian cancer cells. Neoplasia. (2019) 21:206–15. doi: 10.1016/j.neo.2018.12.005
175. Nakamura K, Sawada K, Miyamoto M, Kinose Y, Yoshimura A, Ishida K, et al. Downregulation of miR-194-5p induces paclitaxel resistance in ovarian cancer cells by altering MDM2 expression. Oncotarget. (2019) 10:673–83. doi: 10.18632/oncotarget.26586
176. Li X, Chen W, Jin Y, Xue R, Su J, Mu Z, et al. miR-142-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochem Pharmacol. (2019) 161:98–112. doi: 10.1016/j.bcp.2019.01.009
177. Zhang P, Zhu J, Zheng Y, Zhang H, Sun H, Gao S. miRNA-574-3p inhibits metastasis and chemoresistance of epithelial ovarian cancer (EOC) by negatively regulating epidermal growth factor receptor (EGFR). Am J Transl Res. (2019) 11:4151–65.
178. Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. (2017) 8:59950–64. doi: 10.18632/oncotarget.19048
179. Dai J, Wei R, Zhang P, Kong B. Overexpression of microRNA-195-5p reduces cisplatin resistance and angiogenesis in ovarian cancer by inhibiting the PSAT1-dependent GSK3beta/beta-catenin signaling pathway. J Transl Med. (2019) 17:190. doi: 10.1186/s12967-019-1932-1
180. Chen X, Dong C, Law PT, Chan MT, Su Z, Wang S, et al. MicroRNA-145 targets TRIM2 and exerts tumor-suppressing functions in epithelial ovarian cancer. Gynecol Oncol. (2015) 139:513–9. doi: 10.1016/j.ygyno.2015.10.008
181. Li J, Li D, Zhang W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol Rep. (2016) 35:1671–9. doi: 10.3892/or.2015.4498
182. Duan S, Dong X, Hai J, Jiang J, Wang W, Yang J, et al. MicroRNA-135a-3p is downregulated and serves as a tumour suppressor in ovarian cancer by targeting CCR2. Biomed Pharmacother. (2018) 107:712–20. doi: 10.1016/j.biopha.2018.08.044
183. Liu X, Yang WT, Zheng PS. Msi1 promotes tumor growth and cell proliferation by targeting cell cycle checkpoint proteins p21, p27 and p53 in cervical carcinomas. Oncotarget. (2014) 5:10870–85. doi: 10.18632/oncotarget.2539
184. Li N, Zhao X, Wang L, Zhang S, Cui M, He J. miR-494 suppresses tumor growth of epithelial ovarian carcinoma by targeting IGF1R. Tumour Biol. (2016) 37:7767–76. doi: 10.1007/s13277-015-4603-8
185. Lee M, Kim EJ, Jeon MJ. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget. (2016) 7:8726–42. doi: 10.18632/oncotarget.6474
186. Kinose Y, Sawada K, Nakamura K, Sawada I, Toda A, Nakatsuka E, et al. The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget. (2015) 6:11342–56. doi: 10.18632/oncotarget.3604
187. Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. (2013) 3:1302–15. doi: 10.1158/2159-8290.CD-13-0159
188. Sowter HM, Ashworth A. BRCA1 and BRCA2 as ovarian cancer susceptibility genes. Carcinogenesis. (2005) 26:1651–6. doi: 10.1093/carcin/bgi136
189. Fu X, Li Y, Alvero A, Li J, Wu Q, Xiao Q, et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget. (2016) 7:80633–54. doi: 10.18632/oncotarget.13017
190. Shi C, Zhang Z. miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma. Tumour Biol. (2016) 37:5437–43. doi: 10.1007/s13277-015-4377-z
191. Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol. (2012) 228:204–15. doi: 10.1002/path.4000
192. Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. (2014) 11:495–502. doi: 10.1038/cmi.2014.30
193. Li P, Sun Y, Liu Q. MicroRNA-340 induces apoptosis and inhibits metastasis of ovarian cancer cells by inactivation of NF-x03BA;B1. Cell Physiol Biochem. (2016) 38:1915–27. doi: 10.1159/000445553
194. Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. (2015) 75:117–22. doi: 10.1016/j.biopha.2015.07.035
195. Xia B, Li H, Yang S, Liu T, Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. (2016) 37:9157–67. doi: 10.1007/s13277-016-4805-8
196. Ohyagi-Hara C, Sawada K, Kamiura S, Tomita Y, Isobe A, Hashimoto K, et al. miR-92a inhibits peritoneal dissemination of ovarian cancer cells by inhibiting integrin alpha5 expression. Am J Pathol. (2013) 182:1876–89. doi: 10.1016/j.ajpath.2013.01.039
197. Wen C, Liu X, Ma H, Zhang W, Li H. miR3383p suppresses tumor growth of ovarian epithelial carcinoma by targeting Runx2. Int J Oncol. (2015) 46:2277–85. doi: 10.3892/ijo.2015.2929
198. Yao M, Fang W, Smart C, Hu Q, Huang S, Alvarez N, et al. CCR2 chemokine receptors enhance growth and cell-cycle progression of breast cancer cells through SRC and PKC activation. Mol Cancer Res. (2019) 17:604–17. doi: 10.1158/1541-7786.MCR-18-0750
199. Jiang H, Qu L, Wang Y, Cong J, Wang W, Yang X. miR-99a promotes proliferation targeting FGFR3 in human epithelial ovarian cancer cells. Biomed Pharmacother. (2014) 68:163–9. doi: 10.1016/j.biopha.2013.12.001
200. Hsu CY, Hsieh TH, Er TK, Chen HS, Tsai CC, Tsai EM. MiR381 regulates cell motility, growth and colony formation through PIK3CA in endometriosisassociated clear cell and endometrioid ovarian cancer. Oncol Rep. (2018) 40:3734–42. doi: 10.3892/or.2018.6779
201. Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. (2009) 276:5537–46. doi: 10.1111/j.1742-4658.2009.07237.x
202. Ge T, Yin M, Yang M, Liu T, Lou G. MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cell Physiol Biochem. (2014) 34:2209–20. doi: 10.1159/000369664
203. Tang W, Jiang Y, Mu X, Xu L, Cheng W, Wang X. MiR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. (2014) 26:1420–6. doi: 10.1016/j.cellsig.2014.03.002
204. Chen L, Zhang F, Sheng XG, Zhang SQ, Chen YT, Liu BW. MicroRNA-106a regulates phosphatase and tensin homologue expression and promotes the proliferation and invasion of ovarian cancer cells. Oncol Rep. (2016) 36:2135–41. doi: 10.3892/or.2016.5010
205. Liu J, Dou Y, Sheng M. Inhibition of microRNA-383 has tumor suppressive effect in human epithelial ovarian cancer through the action on caspase-2 gene. Biomed Pharmacother. (2016) 83:1286–94. doi: 10.1016/j.biopha.2016.07.038
206. Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S, et al. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol. (2016) 143:655–63. doi: 10.1016/j.ygyno.2016.09.010
207. Dai C, Xie Y, Zhuang X, Yuan Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother. (2018) 104:763–70. doi: 10.1016/j.biopha.2018.05.077
208. Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. (2010) 16:1119–28. doi: 10.1158/1078-0432.CCR-09-2642
209. Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, et al. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. (2012) 40:761–74. doi: 10.1093/nar/gkr730
210. Xu B, Lefringhouse J, Liu Z, West D, Baldwin LA, Ou C, et al. Inhibition of the integrin/FAK signaling axis and c-Myc synergistically disrupts ovarian cancer malignancy. Oncogenesis. (2017) 6:e295. doi: 10.1038/oncsis.2016.86
211. Roh JW, Huang J, Hu W, Yang X, Jennings NB, Sehgal V, et al. Biologic effects of platelet-derived growth factor receptor alpha blockade in uterine cancer. Clin Cancer Res. (2014) 20:2740–50. doi: 10.1158/1078-0432.CCR-13-2507
212. Ge G, Zhang W, Niu L, Yan Y, Ren Y, Zou Y. miR-215 functions as a tumor suppressor in epithelial ovarian cancer through regulation of the X-chromosome-linked inhibitor of apoptosis. Oncol Rep. (2016) 35:1816–22. doi: 10.3892/or.2015.4482
213. Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. (2010) 10:165–80. doi: 10.1038/nrc2806
214. Tan G, Cao X, Dai Q, Zhang B, Huang J, Xiong S, et al. A novel role for microRNA-129-5p in inhibiting ovarian cancer cell proliferation and survival via direct suppression of transcriptional co-activators YAP and TAZ. Oncotarget. (2015) 6:8676–86. doi: 10.18632/oncotarget.3254
215. Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. (2010) 29:4971–9. doi: 10.1038/onc.2010.233
216. Zhu T, Yuan J, Wang Y, Gong C, Xie Y, Li H. MiR-661 contributed to cell proliferation of human ovarian cancer cells by repressing INPP5J expression. Biomed Pharmacother. (2015) 75:123–8. doi: 10.1016/j.biopha.2015.07.023
217. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. (2017) 17:318–32. doi: 10.1038/nrc.2017.8
218. Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. (2012) 27:594–8. doi: 10.3892/or.2011.1530
219. Zhang Z, Zhang L, Wang B, Wei R, Wang Y, Wan J, et al. MiR-337-3p suppresses proliferation of epithelial ovarian cancer by targeting PIK3CA and PIK3CB. Cancer Lett. (2020) 469:54–67. doi: 10.1016/j.canlet.2019.10.021
220. Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, et al. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci. (2011) 124(Pt 3):359–68. doi: 10.1242/jcs.072223
221. Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med. (2016) 94:1313–26. doi: 10.1007/s00109-016-1475-3
222. Obexer P, Ausserlechner MJ. X-linked inhibitor of apoptosis protein - a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front Oncol. (2014) 4:197. doi: 10.3389/fonc.2014.00197
223. Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. (2014) 35:12339–44. doi: 10.1007/s13277-014-2546-0
224. Liu Y, Lin J, Zhai S, Sun C, Xu C, Zhou H, et al. MicroRNA-214 suppresses ovarian cancer by targeting beta-catenin. Cell Physiol Biochem. (2018) 45:1654–62. doi: 10.1159/000487733
225. Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. (2010) 9:3892–903. doi: 10.4161/cc.9.19.13386
226. Wu AH, Huang YL, Zhang LZ, Tian G, Liao QZ, Chen SL. MiR-572 prompted cell proliferation of human ovarian cancer cells by suppressing PPP2R2C expression. Biomed Pharmacother. (2016) 77:92–7. doi: 10.1016/j.biopha.2015.12.005
227. Harrington BS, Annunziata CM. NF-kappaB signaling in ovarian cancer. Cancers. (2019) 11:1182. doi: 10.3390/cancers11081182
228. Wei Z, Liu Y, Wang Y, Zhang Y, Luo Q, Man X, et al. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol. (2016) 33:126. doi: 10.1007/s12032-016-0842-9
229. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. (2010) 10:116–29. doi: 10.1038/nrc2780
Keywords: microRNA, ovarian cancer, proliferation, biomarkers, chemoresistance, diagnosis, prognosis, target genes
Citation: Alshamrani AA (2020) Roles of microRNAs in Ovarian Cancer Tumorigenesis: Two Decades Later, What Have We Learned? Front. Oncol. 10:1084. doi: 10.3389/fonc.2020.01084
Received: 14 March 2020; Accepted: 29 May 2020;
Published: 21 July 2020.
Edited by:
Jian-Jun Wei, Northwestern University, United StatesReviewed by:
Miguel F. Segura, Vall d'Hebron Research Institute (VHIR), SpainDaniela Taverna, University of Turin, Italy
Copyright © 2020 Alshamrani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali A. Alshamrani, aaalshamrani@ksu.edu.sa
 Ali A. Alshamrani
Ali A. Alshamrani