Abstract
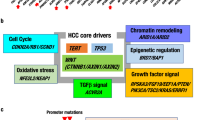
Diverse epidemiological factors are associated with hepatocellular carcinoma (HCC) prevalence in different populations. However, the global landscape of the genetic changes in HCC genomes underpinning different epidemiological and ancestral backgrounds still remains uncharted. Here a collection of data from 503 liver cancer genomes from different populations uncovered 30 candidate driver genes and 11 core pathway modules. Furthermore, a collaboration of two large-scale cancer genome projects comparatively analyzed the trans-ancestry substitution signatures in 608 liver cancer cases and identified unique mutational signatures that predominantly contribute to Asian cases. This work elucidates previously unexplored ancestry-associated mutational processes in HCC development. A combination of hotspot TERT promoter mutation, TERT focal amplification and viral genome integration occurs in more than 68% of cases, implicating TERT as a central and ancestry-independent node of hepatocarcinogenesis. Newly identified alterations in genes encoding metabolic enzymes, chromatin remodelers and a high proportion of mTOR pathway activations offer potential therapeutic and diagnostic opportunities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Forner, A., Llovet, J.M. & Bruix, J. Hepatocellular carcinoma. Lancet 379, 1245–1255 (2012).
El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–1273 (2012).
Yu, J., Shen, J., Sun, T.T., Zhang, X. & Wong, N. Obesity, insulin resistance, NASH and hepatocellular carcinoma. Semin. Cancer Biol. 23, 483–491 (2013).
Augustine, M.M. & Fong, Y. Epidemiology and risk factors of biliary tract and primary liver tumors. Surg. Oncol. Clin. N. Am. 23, 171–188 (2014).
Tanaka, K., Sakai, H., Hashizume, M. & Hirohata, T. Serum testosterone:estradiol ratio and the development of hepatocellular carcinoma among male cirrhotic patients. Cancer Res. 60, 5106–5110 (2000).
International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Wang, K. et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology 58, 706–717 (2013).
Sung, W.K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 44, 765–769 (2012).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764 (2012).
Killela, P.J. et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 110, 6021–6026 (2013).
Nault, J.C. et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 2218 (2013).
Li, Y. & Tergaonkar, V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 74, 1639–1644 (2014).
Heaphy, C.M. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011).
Hoffmeyer, K. et al. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336, 1549–1554 (2012).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Li, M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Guichard, C. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698 (2012).
Kan, Z. et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 23, 1422–1433 (2013).
Tetsu, O. & McCormick, F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999).
Pai, R. et al. Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating β-catenin signaling. Cancer Res. 68, 5086–5095 (2008).
Motohashi, H. & Yamamoto, M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549–557 (2004).
Zhang, D.D., Lo, S.C., Cross, J.V., Templeton, D.J. & Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941–10953 (2004).
Song, L.N. & Gelmann, E.P. Silencing mediator for retinoid and thyroid hormone receptor and nuclear receptor corepressor attenuate transcriptional activation by the β-catenin–TCF4 complex. J. Biol. Chem. 283, 25988–25999 (2008).
Eissenberg, J.C., Wong, M. & Chrivia, J.C. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the Notch signaling pathway. Mol. Cell. Biol. 25, 6559–6569 (2005).
Monroy, M.A. et al. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor–mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 17, 2519–2528 (2003).
Hood, R.L. et al. Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am. J. Hum. Genet. 90, 308–313 (2012).
Nelson, R.A. et al. Floating-Harbor syndrome and intramedullary spinal cord ganglioglioma: case report and observations from the literature. Am. J. Med. Genet. A. 149A, 2265–2269 (2009).
Iyer, G. et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 338, 221 (2012).
Pleasance, E.D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Pleasance, E.D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Poon, S.L. et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med. 5, 197ra101 (2013).
Goedde, H.W. et al. Population genetic studies on aldehyde dehydrogenase isozyme deficiency and alcohol sensitivity. Am. J. Hum. Genet. 35, 769–772 (1983).
Keng, V.W. et al. Sex bias occurrence of hepatocellular carcinoma in Poly7 molecular subclass is associated with EGFR. Hepatology 57, 120–130 (2013).
Zhang, H. et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat. Genet. 42, 755–758 (2010).
Kumar, V. et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 43, 455–458 (2011).
Harley, C.B. Telomerase and cancer therapeutics. Nat. Rev. Cancer 8, 167–179 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Koboldt, D.C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2010).
Sanchez-Garcia, F., Akavia, U.D., Mozes, E. & Pe'er, D. JISTIC: identification of significant targets in cancer. BMC Bioinformatics 11, 189 (2010).
Cox, D.R. & Oakes, D. Analysis of Survival Data (Chapman & Hall/CRC, Boca Raton, FL, 1984).
Acknowledgements
This study was supported by Grants-in-Aid from the Ministry of Health, Labour and Welfare of Japan for the third-term Comprehensive 10-Year Strategy for Cancer Control, grants from the US National Human Genome Research Institute (NHGRI; 5U54HG003273) and National Cancer Institute (NCI; HHSN261201000053C and P30 CA125123), the Program for Promotion of Fundamental Studies in Health Sciences from the National Institute of Biomedical Innovation (NIBIO, Japan) and the National Cancer Center Research and Development Funds (23-A-8, Japan). The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund, Japan. The supercomputing resource SHIROKANE was provided by the Human Genome Center at the University of Tokyo (http://sc.hgc.jp/shirokane.html).
Author information
Authors and Affiliations
Contributions
Study design: Y.T., K.T., K.R.C., H.U., M.K., D.A.W., H.A. and T.S. Sequencing data generation: K.T., D.M.M., F.H., H. Doddapaneni, H. Dinh, Y.A., K.G., K.W., M.-C.G., T.U., S.O., N.O., M.W. and Y.Z. Data analysis: Y.T., K.T., K.R.C., H.U., M.K., S.T., L.A.D., B.L.S., E.S., S.Y., H.N., M.L., N.H., K.W., K.G., M.D., G.N., D.A.W. and T.S. Statistical analysis: Y.T., K.R.C., H.U., K.T., C.J.C., M.K., S.T. and S.Y. Molecular analysis: Y.A. and T.S. Sample acquisition and clinical data collection: M.-C.G., K.S., Y.M., J.A.G., H.O., A.H., J.S., R.C., J.G., S.I., M.T., T.O., N.K., T.K., T.T. and M.F. Manuscript writing: Y.T., K.T., K.R.C., H.U., C.J.C., L.A.D., B.L.S., M.K., D.A.W., H.A. and T.S. Project oversight: D.A.W., R.A.G., H.A. and T.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–35, Supplementary Tables 1, 2, 4–6 and 13–32. (PDF 8833 kb)
Supplementary Tables 3 and 7–12
Supplementary Tables 3 and 7–12. (XLSX 4247 kb)
Rights and permissions
About this article
Cite this article
Totoki, Y., Tatsuno, K., Covington, K. et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 46, 1267–1273 (2014). https://doi.org/10.1038/ng.3126
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3126
This article is cited by
-
Major genomic mutations driving hepatocellular carcinoma
Genome Instability & Disease (2023)
-
Liquid biopsy using cell-free DNA in the early diagnosis of hepatocellular carcinoma
Investigational New Drugs (2023)
-
Fragment length profiles of cancer mutations enhance detection of circulating tumor DNA in patients with early-stage hepatocellular carcinoma
BMC Cancer (2023)
-
Green tea-derived theabrownin induces cellular senescence and apoptosis of hepatocellular carcinoma through p53 signaling activation and bypassed JNK signaling suppression
Cancer Cell International (2022)
-
Demethylation at enhancer upregulates MCM2 and NUP37 expression predicting poor survival in hepatocellular carcinoma patients
Journal of Translational Medicine (2022)