Abstract
The methylation of cytosine and subsequent oxidation constitutes a fundamental epigenetic modification in mammalian genomes, and its abnormalities are intimately coupled to various pathogenic processes including cancer development. Enzymes of the Ten–eleven translocation (TET) family catalyze the stepwise oxidation of 5-methylcytosine in DNA to 5-hydroxymethylcytosine and further oxidation products. These oxidized 5-methylcytosine derivatives represent intermediates in the reversal of cytosine methylation, and also serve as stable epigenetic modifications that exert distinctive regulatory roles. It is becoming increasingly obvious that TET proteins and their catalytic products are key regulators of embryonic development, stem cell functions and lineage specification. Over the past several years, the function of TET proteins as a barrier between normal and malignant states has been extensively investigated. Dysregulation of TET protein expression or function is commonly observed in a wide range of cancers. Notably, TET loss-of-function is causally related to the onset and progression of hematologic malignancy in vivo. In this review, we focus on recent advances in the mechanistic understanding of DNA methylation–demethylation dynamics, and their potential regulatory functions in cellular differentiation and oncogenic transformation.
Similar content being viewed by others
Introduction
Eukaryotic DNA is tightly packaged into a highly organized chromatin structure with the assistance of special proteins called histones.1 Approximately 146 base pairs of DNA are wrapped around a histone octamer that consists of two copies of four core histones (H2A, H2B, H3 and H4) to form the nucleosome, the smallest unit of chromatin. Nucleosomes are then linked by another histone protein called histone H1, followed by further compaction into a higher-order structure that makes up chromosomes. The amino-terminal tails of the core histone proteins are frequently subject to multivalent post-translational modifications, such as acetylation, phosphorylation, methylation, sumoylation and ubiquitylation, altering the degree of local chromatin condensation and accessibility of genetic loci to the cellular machinery that dynamically modulates chromatin architecture and gene expression.
In addition to these histone modifications, a methyl group can be covalently attached to the carbon-5 position of a cytosine (C) in DNA to form 5-methylcytosine (5mC). This process, called ‘DNA methylation’, is a type of epigenetic mechanism that influences transcription, X-chromosome inactivation, suppression of mobile genetic elements and genomic imprinting.2 Recent studies have demonstrated that adenines in the mammalian genome are also methylated to produce N6-methyladenine, but in this review, DNA methylation refers to only cytosine methylation.3
In most mammalian genomes, cytosine methylation occurs almost exclusively in the context of palindromic CpG dinucleotides.4, 5 Typically, cytosines in both strands of a DNA duplex are methylated symmetrically. CpG methylation is catalyzed by a family of DNA methyltransferases (DNMTs), which are classified into two large categories.6 During early embryogenesis, DNMT3A and DNMT3B initially deposit methylation marks on unmethylated CpG, and thus are classified as de novo methyltransferases. Then, DNMT1, a maintenance methyltransferase, is largely responsible for the post-replicative inheritance of pre-existing methylation marks. During semi-conservative DNA replication, the ubiquitin-like plant homeodomain and RING finger domain 1 (UHRF1) preferentially recognizes CpGs in the hemi-methylated DNA via its SET and RING-associated (SRA) domain, and recruits DNMT1 to restore parental methylation patterns on the nascent strand.7, 8, 9, 10, 11 Therefore, the absence of DNMT1/UHRF1 can lead to the progressive dilution of cytosine methylation during successive rounds of DNA replication, a process called ‘passive demethylation’. In addition, DNA demethylation can also take place in a replication-independent manner via the combined action of various enzymes, as described later.
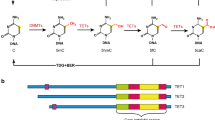
It has long been considered that 5mC is a stably inherited epigenetic modification. However, a subset of 5mCs in the genome are epigenetically unstable and can be further modified enzymatically. Analyses of TET enzyme function have revealed that cytosine in DNA does not exist in a binary modification status (C versus 5mC) as previously believed, but it could adopt one of five different states.12 In the early 2000s, the TET1 gene was first cloned as a fusion partner of mixed-lineage leukemia (MLL) H3K4 methyltransferase (also known as KMT2A) in a handful of acute myeloid and lymphocytic leukemia patients harboring the chromosomal rearrangement t(10;11)(q22;q23).13, 14 By a homology search, additional TET genes, TET2 and TET3, were also identified. However, TET protein function has only recently been determined. TET1 was identified in a search for mammalian homologs of J-binding protein (JBP) 1 and 2, the Fe(II) and 2-oxoglutarate (2OG)-dependent dioxygenases in Trypanosoma brucei that oxidize thymine in DNA to 5-hydroxymethyluracil (5hmU) during the synthesis of base J.15, 16, 17 TET1 was shown to oxidize 5mC to 5hmC in cells and in vitro. The two cofactors, Fe(II) and 2OG, are indispensable for TET-mediated 5mC oxidation. Subsequent studies have shown that all three TET proteins belong to a family of dioxygenase enzymes and share identical catalytic activity to successively oxidize the methyl group of 5mC, yielding three distinct forms of oxidized methylcytosines (termed ‘oxi-mCs’), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC).18, 19, 20, 21
Dysregulation of DNA methylation is a prominent feature of cancers.22 Recent studies have clearly established that 5mC oxidation is also highly disrupted in most cancer types.23, 24, 25, 26, 27 Numerous studies point to the fundamental roles of the key epigenetic regulators such as DNMTs, TETs and isocitrate dehydrogenase (IDH) enzymes in gene expression, development, cellular development and transformation.28 Despite strenuous efforts over the last decade, the exact mechanism underlying enhanced malignant transformation upon the dysregulation of these factors remains poorly understood. Haematopoietic differentiation and transformation is one of the most extensively studied systems in this regard. Thus, in this review, we focus on the current mechanistic understanding of DNA methylation and demethylation pathways in mammals and its functional implications in cell development and transformation, focusing on the hematopoietic system.
Structural basis for substrate recognition and iterative oxidation by TET proteins
TET proteins contain a carboxyl-terminal core catalytic domain that comprises a conserved cysteine-rich domain and a double-stranded β-helix domain (DSBH, also referred to as a ‘jelly-roll fold’) (Figure 1).16, 17 Within the DSBH domain, there are key catalytic residues that interact with Fe(II) and 2OG. Upon cofactor binding, molecular oxygen oxidizes Fe(II) in the catalytic pocket, thereby inducing the oxidative decarboxylation of 2OG and substrate oxidation.29 A large low-complexity insert is found within the DSBH domain and located at the exterior surface of the catalytic domain (Figure 1). Although the precise function of this insertion remains to be determined, it may have regulatory roles via post-translational modifications, such as glycosylation and phosphorylation.30, 31 A study has shown that the deletion of this insert markedly increases 5hmC production by the TET2 catalytic domain.32 TET proteins also have an additional domain that potentially regulates their chromatin targeting. At the amino-terminal region, TET1 and TET3 have a DNA-binding domain called the CXXC domain, which is composed of two Cys4-type zinc finger motifs.16, 17, 33 Interestingly, the ancestral TET2 gene underwent a chromosomal inversion during evolution; as a result, the segment encoding its CXXC domain was separated from the region encoding the catalytic domain.34 Thus, the ancestral CXXC domain of TET2 is now encoded separately by a neighboring gene, IDAX (also called CXXC4). The CXXC domain of TET proteins (IDAX CXXC domain in the case of TET2) is highly conserved and preferentially associates with unmethylated CpG-containing sequences.34, 35, 36 The presence or absence of the CXXC domain may affect the genomic distribution of TET proteins; Tet1 is preferentially detected at the promoter CpG islands (CGIs) or enhancers in mouse embryonic stem cells (ESCs), particularly at the former, whereas Tet2 is mostly enriched in gene bodies or enhancer regions.37, 38, 39
Domain structure of TET proteins. The carboxyl-terminal core catalytic domain is highly conserved among all TET family members and consists of a DSBH domain and a cysteine (Cys)-rich domain. The Cys-rich domain is comprised of two subdomains and modulates the chromatin targeting of TET proteins. The DSBH domain harbors key catalytic motifs, including the HxD motif, which interacts with Fe(II) and 2OG. A large low-complexity insert is found within the DSBH domain, but its function remains to be defined.
Structural analyses of TET proteins provide significant insights into how TET enzymes recognize their substrates and catalyze iterative oxidation reactions.32, 40, 41, 42 The crystal structure of the TET2 catalytic core domain revealed that two subdomains of the Cys-rich domain wrap around the DSBH domain on which DNA is located.32 Interestingly, two out of three zinc fingers, coordinated by several residues from the Cys-rich and DSBH domains, bring the two domains into close proximity to facilitate the formation of a compact globular structure, creating a unique structure for DNA substrate recognition.32 TET2 specifically recognizes 5mCpG-containing DNAs with no preference for the flanking sequences, consistent with the fact that 5hmC is almost exclusively located in the CpG context throughout the genome.43 This interaction is stabilized by extensive intermolecular hydrogen bonds between key residues of TET2 and 5mCpGs-flanking phosphates in the DNA backbone. Hydrophobic interactions resulting from base-stacking interactions also contribute to the overall stability of the structure. Interestingly, CpG recognition does not depend on the methyl group of 5mC; accordingly, TET proteins could accommodate the formyl and carboxyl groups of highly oxidized 5mC derivatives at the active site.32, 42
Unlike 5mC, the majority (>80%) of oxi-mCs are deposited asymmetrically on a specific CpG site.43, 44 What is the molecular basis for this strand asymmetry? As observed for 5mC recognition by the SRA domain of DNMT enzymes, TET2 also recognizes oxi-mCs using a base-flipping mechanism. Upon TET2 binding to the symmetrically methylated palindromic CpG DNA, only a single oxidized base in one strand is flipped out of the DNA duplex and incorporated into the active site.32, 40 A similar base-flipping mechanism has also been observed in the structure of the Naegleria Tet-like dioxygenase (NgTet1).42
In mouse ESCs, TET enzymes convert ~10% of 5mCs to 5hmCs, and only a subset (1–10%) of 5hmCs are further oxidized to 5fC/5caC. Therefore, 5hmC is about 10- to 100-fold more prevalent than more oxidized bases in the genome.17, 20, 45, 46, 47 This unequal genomic distribution of oxi-mCs might be attributable, at least in part, to TDG/BER-mediated active demethylation because 5fC and 5caC, but not 5hmC, are reverted to unmethylated cytosines (Figure 2). In addition, a fraction of oxi-mCs, mostly 5hmC, may not undergo entire oxidation reactions because TET enzymes differentiate their substrates. Indeed, TET proteins are less active on 5hmC and 5fC than on 5mC in vitro, indicating a substrate preference.20, 40, 42 TET-mediated oxidation tends to occur preferentially in regions with higher chromatin accessibility. What determines whether oxi-mCs are committed to undergoing further oxidation? Notably, all three oxi-mCs are similarly recognized by TET proteins with comparable binding affinity, and adopt almost identical conformations within active sites.40 However, the hydroxymethyl group and formyl group of 5hmC and 5fC, respectively, adopt a more restrained conformation within active sites by forming hydrogen bonds with N-oxalylglycine (NOG, 2OG under physiological conditions) as well as polar groups of the cytosine ring. This structural restriction prevents hydrogen abstraction, the rate-limiting step for TET-mediated oxidation reactions with a concomitant decrease in catalytic efficiency.40 Collectively, the catalytic core of TET proteins has intrinsic properties for efficient CpG recognition, substrate preference and strand biases (Figure 1). Thus, a fraction of 5hmC is less prone to further oxidation and remains as a stable epigenetic mark.
Function of TET proteins in passive and active DNA demethylation. TET proteins iteratively oxidize 5mC to produce oxidized methylcytosines (oxi-mCs), of which 5fC and 5caC are directly excised by the DNA repair enzyme TDG (thymine DNA glycosylase). The resulting abasic sites are eventually replaced with unmethylated cytosines by base excision repair (BER). No mammalian 5mC glycosylases that directly excise 5mC have been reported to date. TET proteins also promote the oxidative demethylation of 5mC in a replication-dependent manner because oxi-mCs tend to interfere with the methylase activity of DNMT1. TET proteins have a distinct preference for their substrates, so many oxi-mCs, mostly 5hmC, are not committed to demethylation pathways and are stable epigenetic modifications.
Considering the capability of TET enzymes to oxidize their substrates in a step-wise manner, differential genomic levels of oxi-mCs also suggest that TET-catalyzed oxidation is not processive, and frequently stalls at the intermediate stages, most likely at 5hmC. TET proteins may associate transiently with specific substrates and detach before completing oxidation to the end product 5caC. Furthermore, there may be a division of labor among distinct TET enzymes. In fact, a recent study has shown that collaborative interplay among TET proteins and transcription factors is required to complete active DNA demethylation in enhancers.48 In mouse ESCs, Tet1 recruits Sall4, which is a strong 5hmC-interacting protein in vitro, to enhancers. Unexpectedly, the Sall4-bound enhancers are substantially depleted of 5hmCs, but significantly enriched for 5caC. Deletion of Sall4 increases 5hmC levels in these regions in a Tet1-dependent manner, suggesting that Tet1 is mainly responsible for the initial oxidation of 5mC to 5hmC. In contrast, Sall4 loss leads to a reduction in 5caC levels and Tet2 occupancy at the Sall4-bound enhancers. Furthermore, depletion of Tet2, but not Tet1, increases 5hmC levels at Sall4-bound sites. These observations suggest that cooperative interactions between Tet1 and Tet2 are coordinated by an oxi-mC-sensing transcription factor to complete stepwise 5mC oxidation at enhancer regions.
Impact of oxidized methylcytosines in DNA methylation and demethylation
DNA methylation is a highly dynamic process. Therefore, it is important to precisely control the generation and erasure of methylation marks to ensure the long-term inheritance of cell type-specific epigenomic memory across generations.49, 50 As mentioned earlier, following DNA replication, hemi-methylated CpG DNAs are transiently formed with only the parental strand containing 5mC, and the original modification patterns are restored by re-methylating cytosines in the newly synthesized DNA strands (by DNMT1) and consecutively re-oxidizing the resulting 5mCs (by TET proteins). If the methylation maintenance machinery becomes non-functional or chromatin accessibility becomes restricted under certain conditions, 5mCs would be passively diluted as cells divide, either globally or locally. TET proteins can also promote this process, but they first oxidize 5mCs to oxi-mCs, which are subsequently diluted to regenerate unmethylated cytosines in a replication-dependent manner.
Compared to maintenance methylation whose molecular mechanism is relatively well defined, it is not clear how 5mC oxidation patterns are restored and faithfully inherited by daughter cells. It has been shown that maintenance methylation re-establishes methylation patterns immediately after DNA replication, but subsequent TET-mediated oxidation occurs relatively slowly at a later time point.51 TET proteins may not simply catalyze the successive oxidation of 5mCs once they are generated by DNMT1/UHRF1, and different mechanisms might be employed to restore patterns of DNA methylation and oxi-mCs during cell division. How might the oxidized 5mC bases affect passive demethylation? Given that oxi-mCs at CpG-containing DNA interfere with the ability of DNMT1 to methylate CpG sites in vitro,52, 53, 54 TET proteins were proposed to promote replication-dependent passive demethylation (Figure 2). If this is the case, TET proteins might be able to induce progressive DNA demethylation even in the presence of active DNMT1/UHRF1, as observed in normal erythropoiesis (Figure 3).55, 56, 57
A model of TET-assisted passive DNA demethylation. The parental DNA methylation patterns are faithfully inherited to daughter cells across generations because the methylation maintenance machinery DNMT1/UHRF1 is targeted to the hemi-methylated DNA after DNA replication and re-methylates cytosine in the newly synthesized strand. Upon chromatin reorganization at certain genetic loci, such as enhancers, in response to cellular signals, TET proteins and BER components might become more accessible. As a result, a fraction of 5mC may undergo stepwise oxidation. After replication, the resulting DNA contains oxi-mCs only on one strand, which impairs maintenance methylation. Therefore, 5mCs would be passively diluted upon successive cell divisions, even in the presence of functional DNMT1/UHRF1. The impact of de novo DNA methyltransferases in DNA methylation maintenance is not considered here.
Although the result is controversial, the SRA domain of UHRF1 has been shown to recognize 5hmC and 5mC with similar affinity.58 The UHRF2 SRA domain also preferentially recognizes 5hmC.59, 60 As UHRF1 is an obligate partner protein of DNMT1, these results suggest that 5hmC could promote methylation maintenance by facilitating the recruitment of DNMT1 to hemi-hydroxymethylated DNA. Moreover, DNMT3A and DNMT3B, originally known as de novo DNA methyltransferases, are also required for DNA methylation maintenance in somatic cells,61 and they display comparable methylase activity on 5mC- and oxi-mC-containing DNA in vitro, with 5fC increasing methylation efficiency most markedly.53, 54, 62 Thus, further studies are required to elucidate the precise roles of oxi-mCs in the maintenance of DNA methylation.
In addition to passive dilution, 5mCs can also be removed enzymatically by a replication-independent mechanism, a process called ‘active DNA demethylation’ (Figure 2).12, 25, 26, 49 In plants, active demethylation depends on DEMETER and REPRESSOR of SILENCING 1, which are well-characterized 5mC DNA glycosylases that directly excise 5mC to initiate base excision repair (BER). However, no orthologs with similar activities have been identified in mammals. The DNA repair protein thymine DNA glycosylase (TDG), which belongs to the uracil DNA glycosylase superfamily, was a strong candidate owing to its ability to remove the pyrimidine base from a T:G mismatch that arises from the deamination of 5mC.63 However, given the preference of activation-induced deaminase (AID)/APOBEC deaminases for single-stranded DNA and unmethylated cytosine over modified bases, this pathway may play a marginal role.64 Notably, TDG specifically recognizes 5fC and 5caC, but not 5mC and 5hmC, which normally base-pair with guanine, and it shows robust in vitro base excision activity.18, 60, 65, 66 TDG harbors a binding pocket that specifically accommodates these oxidized bases.66 Mechanistically, 5fC and 5caC were shown to destabilize the covalent bond that links them to sugar, making the glycosidic bond more susceptible to cleavage by TDG.67, 68
It is now clear that 5mC in mammalian genomes can be removed by a two-step process (Figure 2). TET proteins first oxidize 5mCs to form oxi-mCs, and TDG subsequently excises the highly oxidized bases 5fC and 5caC.18, 65 This excision reaction results in abasic sites that are eventually repaired by the BER pathway to restore unmodified cytosines. In line with this, the knockdown of Tdg in mouse ESCs leads to a 5- to 10-fold increase in the levels of genomic 5fC/5caC, whereas its overexpression in HEK293T cells markedly diminishes the levels of TET-generated 5fC/5caC.18, 44, 64, 69, 70, 71, 72, 73 Interestingly, vitamin C treatment leads to a significant increase in the levels of 5fC and 5caC, consistent with its function in stimulating the catalytic activity of Tet enzymes.44, 74, 75, 76, 77, 78 In line with its profound role in demethylation, Tdg is essential for embryonic development, as evidenced by mice with Tdg deficiency79, 80 or the expression of mutant Tdg lacking glycosylase activity80, which exhibit developmental defects and embryonic lethality, possibly by impairing the disappearance of 5fC and 5caC. Other studies have shown that 5hmU can be generated as a result of either deamination by AID/APOBEC80, 81 or direct oxidation by TET enzymes,82 followed by TDG-mediated BER. Furthermore, DNMTs could directly catalyze dehydroxymethylation83, 84 and the cell lysate of ESCs exhibits 5caC decarboxylase activity.85 These pathways need to be further characterized in vivo.
Genomic landscape of cytosine methylation and its oxidation products
In mammalian genomes, ~4–5% of all cytosines in the CpG context are methylated to yield 5mC. The methylation frequency at individual CpG sites typically displays a bimodal distribution. In general, the majority (70–80%) of CpG sites within genic and intergenic regions are highly methylated, whereas a small fraction (<20%) that includes promoter CGIs and distal regulatory elements, such as enhancers, is notably depleted of methylation.5, 86, 87, 88, 89 Interestingly, non-CpG methylation is prevalent in ESCs, neuronal precursor cells and ectoderm-derived tissues, such as the cerebellum, cortex and olfactory bulb.5, 89 Cancer cells display highly dysregulated DNA methylation profiles characterized by global hypomethylation, which presumably impairs genome integrity, in conjunction with localized hypermethylation of promoter CGIs associated with aberrant expression of tumor suppressor genes or repair genes.90, 91, 92 However, recent technological advances have enabled the precise mapping of individual cytosine derivatives at single-base resolution, and these analyses have suggested that tumorigenesis is more highly associated with the genome-wide loss of 5hmC than 5mC.93
Interestingly, the global level of cytosine methylation across various human and murine tissues is remarkably similar.88, 89, 94 However, some CpG sites (7–20%) in the mouse epigenome are differentially methylated among cell types; they are mostly hypomethylated in a tissue-specific manner.86, 87, 88, 89 Most of these regions represent the small, evolutionarily conserved, distal cis-regulatory elements marked with H3K4me1, H3K27ac and p300 occupancy, and show significant enrichment of tissue-specific transcription factor binding sites, indicating that they include active enhancers.86, 87, 88, 89 Intriguingly, transcription factor binding is necessary and sufficient to reduce methylation levels in these regions. In particular, cell type-specific transcription factors could locally modify these regions during differentiation, inducing dynamic changes in the expression of the neighboring genes.87
Genome-wide mapping analyses have shown that 5hmC is also strongly enriched in hypomethylated distal regulatory elements, such as enhancers.39, 87, 95 Base-resolution DNA methylome mapping has revealed that Tet deficiency leads to more hypomethylated sites than hypermethylated sites in ESCs.43 Extensive DNA hypermethylation typically occurs in distal enhancer regions that are associated with enhancer-related histone modifications (H3K4me1 and H3K27ac), increased DNase I hypersensitivity, and occupancy by transcription factors and a histone-modifying complex. On the other hand, hypomethylated regions are randomly distributed throughout the genome. Notably, the majority of hypermethylated regions overlap significantly with regions enriched with 5fC and 5caC observed in the Tdg knockdown ESCs, suggesting that Tet-mediated demethylation mainly occurs in these regions. Changes in DNA methylation levels differentially influence the transcription of neighboring genes.95 For example, Tet loss inhibits recruitment of Kap1 to the chromatin and induces derepression of most two-cell embryo (2C)-specific genes such as Zscan4. As expected based on the known function of Zscan4 in telomerase-independent telomere elongation, telomere length is elongated in Tet-deficient ESCs.
On the basis of genome-wide profiling, 5fC and 5caC mostly reside in the distal regulatory elements, including the active/poised enhancers, CTCF-bound insulators, active/poised promoters, and gene bodies of actively transcribed genes.44, 69, 70, 71, 72, 96 Combined with Tdg depletion, these studies have enabled assessments of the dynamics and regulatory mechanisms of active DNA demethylation pathways. Interestingly, 5fC/5caC and 5hmC largely exist at distinct CpGs, and 5fC and 5caC frequently do not overlap at individual CpGs. There are about three times more CpGs modified with 5hmC alone than in association with 5fC/5caC,44 indicating that TET/TDG-mediated active demethylation preferentially stops at the 5hmC step and accordingly the majority of 5hmCs could exist as stable marks. Furthermore, a considerable fraction of 5fC/5caC peaks are found in distal regulatory elements with relatively higher chromatin accessibility, suggesting that the catalytic processivity of TET enzymes is regulated by the local chromatin environment. Interestingly, like 5hmC, most of the 5fC/5aC are asymmetrically modified,47 demonstrating that active DNA demethylation activity targets palindromic CpGs asymmetrically, consistent with the asymmetric base-flipping model.
Oxidized 5-methylcytosine derivatives as distinct epigenetic marks
Oxi-mCs are detectable in most tissues, but their levels are relatively very low compared to those of other bases and highly variable across cell types. 5hmC is most prevalent in ESCs, Purkinje neurons and the brain.45, 94, 97, 98, 99 As discussed, a significant amount of 5hmC is maintained as stable, demethylation-independent bases and can exert independent epigenetic roles.25, 26, 47, 51 The presence of oxi-mCs in DNA influences its physical properties. For example, 5hmC increases the thermodynamic stability of a DNA double helix.100 When 5fC is incorporated into DNA, it induces alterations of the local DNA structure and influences the accessibility of DNA-binding proteins, presumably by altering the degree of DNA supercoiling and packaging. Furthermore, RNA polymerase II specifically recognizes 5caC and 5fC and forms hydrogen bonds with the 5-carboxyl or 5-carbonyl groups of 5caC or 5fC, respectively. As a result, RNA polymerase II is transiently stalled, thereby delaying transcription elongation on gene bodies.101, 102 Moreover, individual oxi-mCs were shown to be specifically recognized by numerous cellular proteins, called ‘oxi-mC readers’, which can differentiate the distinct chemical modification status of oxi-mCs.60, 103, 104, 105, 106 By altering the modification status of different cytosine derivatives, cells might be able to selectively control the chromatin association and dissociation of these cellular proteins. For instance, the transcription factor Wilms tumor 1 binds preferentially to unmethylated or methylated DNA, but binds less efficiently when its cognate binding site contains oxi-mCs. In addition, TET proteins also interact with diverse cellular proteins that potentially affect its chromatin targeting and steady-state levels, as reviewed elsewhere.25, 29
TET proteins in hematologic cancers
TET2 is frequently mutated in a wide spectrum of myeloid malignancies, including ~20% of myelodysplastic syndrome (MDS), 20% of myeloproliferative neoplasms (MPN), 50% of chronic myelomonocytic leukemia (CMML), and 20% of acute myeloid leukemia (AML), reviewed elsewhere.23, 25, 27 TET2 mutations are associated with aberrant DNA methylation patterns in myeloid malignancies. TET2 deletion and mutations are mostly heterozygous and are considered an early event in the pathogenesis of myeloid malignancies. Most of the leukemia-associated TET2 missense mutations are inactivating mutations that inhibit or abolish the catalytic activity of TET2 in vitro and in vivo.21 These mutations may impair the interaction of Fe(II) and 2OG at the active site or affect the structural integrity of the catalytic core domain. Furthermore, TET2 was shown to be monoubiquitylated by the CRL4VprBP E3 ligase, which promotes the chromatin binding of TET2.107 Interestingly, leukemia-associated TET2 mutations are frequently targeted to the residues that are directly ubiquitylated or required for associations with the E3 ligase.
Early studies using hematopoietic stem/progenitor cells (HSPCs) from MPN patients bearing TET2 mutations108 or HSPCs in which Tet2 expression was knocked-down109, 110 have shown that Tet2 inactivation induces a developmental bias toward myeloid lineages at the expense of other lineages. Overall, various Tet2 loss-of-function mouse models exhibit very similar phenotypes, including augmented HSC expansion, increased repopulating capacity of HSCs, and skewed differentiation toward the myeloid lineage.23, 24, 25, 26 Some strains of Tet2-deficient mice, including those containing a homozygous or heterozygous deletion of Tet2, developed myeloid malignancies, indicating a causal relationship between Tet2 loss-of-function and myeloid transformation. Notably, Tet2 deletion in the more highly differentiated myeloid cells compared with HSPCs is not capable of inducing leukemogenesis, and only wild type, but not catalytically inactive Tet2, could rescue the leukemogenic phenotypes in Tet2-deficient mice, suggesting that the catalytic activity of Tet2 is required to suppress myeloid transformation.111 Consistent with recurrent TET2 mutations in a subset of lymphoid malignancies, T-cell lymphoma with follicular helper T-cell-like phenotypes has also been observed in some Tet2-deficient mice.112 These results collectively suggest that TET2 functions as a bona fide tumor suppressor in hematological malignancies. However, it appears that Tet2 deletion/mutation alone is not enough to drive full-blown leukemia. Thus, TET2 dysregulation may contribute to the induction of a pre-leukemic condition. The acquisition of additional mutations may then drive the development of full-blown malignancy. Supporting this hypothesis, Tet2 deficiency has synergistic effects with various leukemia-related mutations that commonly co-exist with TET2 mutations in patients. Depending on the types of second mutations, the fate of leukemic cells could diversify and the disease latency is markedly shortened.113
TET1 also has a regulatory role in hematopoietic transformation. Interestingly, TET1 seems to exert context-dependent effect. TET1 is a direct transcriptional target of MLL fusion proteins and activates the expression of its downstream oncogenic targets to promote leukemogenesis, suggesting its oncogenic roles in MLL-rearranged leukemia.114 In contrast, the loss of Tet1 in mice promotes the development of B-cell lymphoma resembling follicular lymphoma and diffuse large B-cell lymphoma, albeit with a long latency,115 suggesting its tumor suppressor function in lymphomagenesis. In non-Hodgkin B-cell lymphoma (B-NHL), TET1 expression is suppressed at the transcriptional level via promoter CpG methylation. Tet1 deficiency leads to an enhanced serial replating capacity of HSPCs, augmented HSC self-renewal and repopulating capacity, and the accumulation of DNA damage. Tet1 loss also induces developmental bias toward the B-cell lineage.
Tet3 deficiency in mouse HSCs does not show any overt hematopoietic phenotypes, except for the expansion of HSPCs.25 However, Tet2 and Tet3 are highly expressed in the hematopoietic system, suggesting that Tet2 and Tet3 play redundant roles in the regulation of normal hematopoiesis and oncogenesis.116 As expected, the combined loss of Tet2 and Tet3 markedly impairs 5hmC production in hematopoietic cells, suggesting that they are the major 5mC oxidases in the hematopoietic system. Remarkably, the dual loss of Tet2 and Tet3 rapidly induces the development of highly aggressive, fully penetrant and cell-autonomous myeloid leukemia in mice. In Tet2/Tet3 double-deficient HSPCs, the myeloid lineage genes are significantly upregulated, whereas lymphoid and erythroid lineage genes are strongly downregulated. These altered gene expression patterns are associated with myeloid skewing. The double deficiency leads to a mild but consistent increase in DNA methylation, but this altered DNA methylation only has a mild relationship to gene expression levels. Furthermore, upon the loss of Tet2 and Tet3, DNA damage progressively accumulates, suggesting that TET proteins also play significant roles in maintaining genomic integrity.
In addition to myeloid cancers, TET2 mutations are also found in lymphoid cancers, including ~2% of Hodgkin’s lymphoma and 10% of T-cell lymphoma cases. Furthermore, TET1 expression is significantly downregulated in acute B-lymphocytic leukemia. Because both TET1 and TET2 are frequently downregulated in acute B-lymphocytic leukemia, the impact of the simultaneous deletion of both genes on hematopoietic development has been tested.117 Surprisingly, Tet1/Tet2 double knockout mice show significant decreases in the frequency of myeloid malignancies and have a strikingly improved survival rate compared to that of Tet2-deficient mice. Even haplo-insufficiency of Tet1 is sufficient to induce these phenotypes in Tet2-deficient mice. Furthermore, the double knockout mice mainly develop transplantable, lethal B-acute lymphoblastic leukemia-like malignancies associated with the clonal expansion of B cells, extensive lymphocyte infiltration into the bone marrow, spleen and liver, spleno-hepatomegaly, and enlarged lymph nodes.
Additional major epigenetic factors in hematopoietic cancers
IDH enzymes
Recent studies suggest that an altered metabolic status is closely linked to cellular transformation because many key enzymes implicated in tumor suppression consume various metabolites as cofactors. TET proteins require 2OG to catalyze 5mC oxidation. 2OG is mainly produced by IDH enzymes in the TCA cycle that catalyze the oxidative decarboxylation of isocitrate (Figure 4). Interestingly, recurrent heterozygous mutations in IDH1 and IDH2 genes have been detected in a majority of glioblastomas and various hematopoietic malignancies, including MDS, MPN and AML.25 IDH mutations are almost exclusively targeted to specific mutational hotspots (R132 in IDH1 and R140 and R172 in IDH2) and confer a neomorphic ability to reduce 2OG to 2-hydroxyglutarate (2HG) (Figure 4).118 Thus, patients with IDH mutations show elevated levels of 2HG. In addition, inactivating mutations frequently arise in other genes that encode additional metabolic enzymes. For example, mutations in succinate dehydrogenase (SDH) and fumarate hydratase (FH) lead to the accumulation of succinate and fumarate. Interestingly, the structures of 2HG, succinate and fumarate are very similar to that of 2OG. Accordingly, they can compete with 2OG to inhibit 2OG-dependent dioxygenases, including TETs and JmjC-domain-containing histone demethylases, causing an increase in histone and DNA methylation (Figure 4). As a way of targeting mutant IDH enzymes to treat cancers, specific inhibitors that interfere with 2HG production by mutant IDH enzymes have been developed and they were shown to have clinical efficacy against gliomas in vitro and in vivo.119
TET protein as a linker between metabolism and epigenetic regulation. During the tricarboxylic acid (TCA) cycle, IDH enzymes catalyze the oxidative decarboxylation of isocitrate to generate 2-oxoglutarate (2OG), an essential co-substrate that TET enzymes require to oxidize their substrates. Mutations in the IDH1 gene increase the binding affinity for NADPH relative to isocitrate and NADP+; thus, the resulting mutant enzymes acquire neomorphic activity to reduce 2OG to 2-hydroxyglutarate (2HG). Owing to the structural similarity, 2HG can function as a competitive inhibitor of TET enzymes.
To characterize the in vivo function of IDH mutations, several mouse models, including those expressing mutant IDH1 or IDH2, have been generated.120, 121 Although the expression of mutant IDH in mice leads to abnormal hematopoietic phenotypes, the mice were not the exact phenocopies of those with Tet2 deficiency. For example, IDH mutations do not significantly affect myeloid differentiation and the repopulating capacity of HSCs, which are consistently observed in various Tet2-deficient mouse models. Furthermore, no leukemogenesis has been observed in any of these mouse models. Thus, these results suggest that IDH mutations alone contribute to pre-leukemic conditions, and full-blown leukemia develops via the gain of additional mutations. Interestingly, genetic or pharmacological suppression of mutant IDH proteins could promote the differentiation of leukemic cells and significantly ameliorate the pathogenic features, suggesting the requirement of 2HG in the maintenance of leukemic cells.121, 122
DNMT3A
During hematopoiesis, DNA methylation pattern is dynamically regulated.123, 124 Individual DNMTs have been shown to be critical for HSC self-renewal, normal hematopoietic differentiation, lineage specification and suppression of malignant transformation.24 Among them, the de novo DNA methyltransferase DNMT3A has gained much attention. In mice, the loss of Dnmt3a in HSCs augments HSC self-renewal and impairs differentiation over serial transplantation,125 which was further enhanced by the additional loss of Dnmt3b.126 Dnmt3a-deficient HSCs show aberrant DNA methylation patterns, but changes in DNA methylation are not strongly correlated with alterations in gene expression levels. HSCs doubly deficient in Dnmt3a and Dnmt3b have large hypomethylated regions in the CGI shore in the β-catenin (Ctnnb1) promoter, which transcriptionally upregulates β-catenin and its downstream target genes to block HSC differentiation.
DNMT3A is also frequently mutated in a wide range of hematopoietic malignancies including AMLs (20–30%), MDS (10–15%), and MPN (~8%), and DNMT3A mutations are generally correlated with poor prognosis.127 These mutations are typically heterozygous and target a specific residue, arginine 882, in the catalytic domain. The DNMT3AR882H mutant has a dominant negative effect. The expression of the DNMT3AR882H mutant or the deletion of Dnmt3a in mice leads to the development of a wide spectrum of myeloid and lymphoid malignancies resembling MDS, MPN, CMML, AML and acute lymphoblastic leukemia although the disease latency is very long. Similar to TET2 mutations, DNMT3A mutations are considered an early event that are introduced in HSCs, inducing a pre-leukemic condition, and a Dnmt3a deficiency cooperates with MLL-AF9, Flt3-ITD, and other mutations such as c-Kit, Kras and Npm1 mutations to promote oncogenic transformation toward a diverse spectrum of malignancies.
DNMT3A mutations frequently co-exist with TET2 mutations in lymphoma and leukemia. Mutations in both genes are expected to modulate DNA methylation patterns in opposite directions; the former leads to global hypomethylation in general, whereas the latter leads to hypermethylation. However, in studies involving Dnmt3a- or Tet2-deficient mice, the pathological outcomes are very similar. Because TET2 consumes 5mC generated by DNMT3A, both mutations would result in the same end result at the molecular level, that is, a loss of oxi-mCs. A recent study has shown that compared to single deletions, the combined deletion of Dnmt3a and Tet2 in mice further augments the accumulation and repopulating capacity of HSPCs, and accelerates the development of hematologic malignancy, including B-cell and T-cell lymphomas,128 similar to the DNMT3AR882H in the Tet2-deficient background.129 The dual loss of both enzymes results in the downregulation of HSC-specific genes and derepression of lineage-specific genes. For example, Dnmt3a and Tet2 collaborate to prevent the activation of Klf1 and Epor. These genes are known as erythroid lineage genes, but erythropoiesis is paradoxically blocked in the double knockout mice, resulting in anemia. Interestingly, these genes promote the self-renewal of double-knockout HSPCs in vitro. Further studies are required to precisely assess whether the loss of 5mC, oxi-mC or both contributes to malignant hematopoiesis.
Conclusions and perspective
DNA methylation plays pivotal regulatory roles in diverse cellular processes, such as transcription and genome integrity, and its aberrations influence mammalian development and cancer development. TET proteins directly modulate the DNA methylation landscape by successively oxidizing 5mCs. TET loss-of-function is commonly observed in various cancers, including hematopoietic and non-hematopoietic cancers, and studies of various mouse models have clearly shown that it is causally related to the pathogenesis of hematologic cancers. Notably, the re-introduction of wild-type Tet activity into Tet-deficient HSPCs fully rescues the leukemogenic phenotypes in mice. Similar tumor-suppressor functions are anticipated for the wide spectrum of solid cancers. Therefore, the restoration of TET expression or function in cancers will have an immense clinical impact. In this regard, it is noteworthy that the combined treatment of DNMT inhibitors and vitamin C shows a marked effect in restoring TET activity in cancers. Despite vast information on the regulatory function of TET proteins in stem cell maintenance, lineage specification, gene transcription, genomic integrity and oncogenesis, it is still unclear how TETs control normal cell differentiation and malignant transformation. Further studies are required to uncover the exact molecular mechanism underlying accelerated oncogenesis upon TET loss-of-function. Furthermore, it is also necessary to develop tools to precisely manipulate TET function in cancer cells and identify targets for therapeutic intervention and/or preventive measures.
References
Andrews AJ, Luger K . Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys 2011; 40: 99–117.
Smith ZD, Meissner A . DNA methylation: roles in mammalian development. Nat Rev Genet 2013; 14: 204–220.
Iyer LM, Zhang D, Aravind L . Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 2016; 38: 27–40.
Gruenbaum Y, Stein R, Cedar H, Razin A . Methylation of CpG sequences in eukaryotic DNA. FEBS Lett 1981; 124: 67–71.
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462: 315–322.
Goll MG, Bestor TH . Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74: 481–514.
Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE . UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317: 1760–1764.
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450: 908–912.
Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M . Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008; 455: 818–821.
Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X . The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008; 455: 826–829.
Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008; 455: 822–825.
Pastor WA, Aravind L, Rao A . TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14: 341–356.
Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y . LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 2002; 62: 4075–4080.
Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR . TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 2003; 17: 637–641.
Borst P, Sabatini R, Base J . discovery, biosynthesis, and possible functions. Annu Rev Microbiol 2008; 62: 235–251.
Iyer LM, Tahiliani M, Rao A, Aravind L . Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 2009; 8: 1698–1710.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930–935.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333: 1303–1307.
Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y . Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466: 1129–1133.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333: 1300–1303.
Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010; 468: 839–843.
Iacobuzio-Donahue CA . Epigenetic changes in cancer. Annu Rev Pathol 2009; 4: 229–249.
Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I . TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 2011; 9: 193–204.
Ko M, An J, Rao A . DNA methylation and hydroxymethylation in hematologic differentiation and transformation. Curr Opin Cell Biol 2015; 37: 91–101.
Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A . TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 2015; 263: 6–21.
Han JA, An J, Ko M . Functions of TET proteins in hematopoietic transformation. Mol Cells 2015; 38: 925–935.
Huang Y, Rao A . Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet 2014; 30: 464–474.
Guillamot M, Cimmino L, Aifantis I . The impact of DNA methylation in hematopoietic malignancies. Trends Cancer 2016; 2: 70–83.
Shen L, Song CX, He C, Zhang Y . Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem 2014; 83: 585–614.
Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y et al. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 2009; 5: 204–213.
Bauer C, Göbel K, Nagaraj N, Colantuoni C, Wang M, Müller U et al. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). J Biol Chem 2015; 290: 4801–4812.
Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 2013; 155: 1545–1555.
Long HK, Blackledge NP, Klose RJ . ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans 2013; 41: 727–740.
Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013; 497: 122–126.
Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG et al. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res 2010; 20: 1390–1393.
Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 2012; 151: 1200–1213.
Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA 2014; 111: 1361–1366.
Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011; 473: 389–393.
Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 2014; 56: 286–297.
Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 2015; 527: 118–122.
Hashimoto H, Pais JE, Dai N, Corrêa IR Jr, Zhang X, Zheng Y et al. Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA. Nucleic Acids Res 2015; 43: 10713–10721.
Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 2014; 506: 391–395.
Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012; 149: 1368–1380.
Wu H, Wu X, Shen L, Zhang Y . Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol 2014; 32: 1231–1240.
Wu H, Zhang Y . Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol 2015; 22: 656–661.
Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 2011; 50: 7008–7012.
Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S . Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat Chem 2014; 6: 435–440.
Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X et al. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol Cell 2016; 64: 913–925.
Wu H, Zhang Y . Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014; 156: 45–68.
Ooi SK, Bestor TH . The colorful history of active DNA demethylation. Cell 2008; 133: 1145–1148.
Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S . 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 2014; 6: 1049–1055.
Valinluck V, Sowers LC . Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 2007; 67: 946–950.
Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012; 40: 4841–4849.
Ji D, Lin K, Song J, Wang Y . Effects of Tet-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol Biosyst 2014; 10: 1749–1752.
Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M . Global DNA demethylation during mouse erythropoiesis in vivo. Science 2011; 334: 799–802.
Madzo J, Liu H, Rodriguez A, Vasanthakumar A, Sundaravel S, Caces DB et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep 2014; 6: 231–244.
Ruiz MA, Rivers A, Ibanez V, Vaitkus K, Mahmud N, DeSimone J et al. Hydroxymethylcytosine and demethylation of the gamma-globin gene promoter during erythroid differentiation. Epigenetics 2015; 10: 397–407.
Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE 2011; 6: e21306.
Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B et al. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol Cell 2014; 54: 879–886.
Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013; 152: 1146–1159.
Jones PA, Liang G . Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009; 10: 805–811.
Otani J, Kimura H, Sharif J, Endo TA, Mishima Y, Kawakami T et al. Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PLoS ONE 2013; 8: e82961.
Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P . The enigmatic thymine DNA glycosylase. DNA Repair 2007; 6: 489–504.
Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 2012; 8: 751–758.
Maiti A, Drohat AC . Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 2011; 286: 35334–35338.
Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 2012; 8: 328–330.
Williams RT, Wang Y . A density functional theory study on the kinetics and thermodynamics of N-glycosidic bond cleavage in 5-substituted 2′-deoxycytidines. Biochemistry 2012; 51: 6458–6462.
Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC . Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J Am Chem Soc 2006; 128: 12510–12519.
Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 2013; 153: 692–706.
Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 2013; 153: 678–691.
Xia B, Han D, Lu X, Sun Z, Zhou A, Yin Q et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat Methods 2015; 12: 1047–1050.
Lu X, Han D, Zhao BS, Song CX, Zhang LS, Doré LC et al. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res 2015; 25: 386–389.
Neri F, Incarnato D, Krepelova A, Rapelli S, Anselmi F, Parlato C et al. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep 2015; 10: 674–683.
Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013; 500: 222–226.
Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet 2013; 45: 1504–1509.
Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 2010; 6: 71–79.
Minor EA, Court BL, Young JI, Wang G . Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem 2013; 288: 13669–13674.
Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 2013; 135: 10396–10403.
Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 2011; 470: 419–423.
Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011; 146: 67–79.
Guo JU, Su Y, Zhong C, Ming GL, Song H . Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011; 145: 423–434.
Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol 2014; 10: 574–581.
Chen CC, Wang KY, Shen CK . The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem 2012; 287: 33116–33121.
Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S . Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol 2009; 5: 400–402.
Schiesser S, Hackner B, Pfaffeneder T, Müller M, Hagemeier C, Truss M et al. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl 2012; 51: 6516–6520.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770.
Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011; 480: 490–495.
Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013; 500: 477–481.
Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 2013; 45: 1198–1206.
Eden A, Gaudet F, Waghmare A, Jaenisch R . Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003; 300: 455.
Herman JG, Baylin SB . Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349: 2042–2054.
Esteller M . Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159.
Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res 2016; 26: 103–118.
Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 2010; 5: e15367.
Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y . Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 2014; 28: 2103–2119.
Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE . 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 2011; 12: R54.
Kriaucionis S, Heintz N . The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009; 324: 929–930.
Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H . Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 2010; 38: e181.
Song CX, Yi C, He C . Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol 2012; 30: 1107–1116.
Raiber EA, Murat P, Chirgadze DY, Beraldi D, Luisi BF, Balasubramanian S . 5-Formylcytosine alters the structure of the DNA double helix. Nat Struct Mol Biol 2015; 22: 44–49.
Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D . 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol 2012; 19: 831–833.
Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 2015; 523: 621–625.
Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol 2013; 14: R119.
Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X . Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 2014; 28: 2304–2313.
Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N . MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012; 151: 1417–1430.
Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011; 147: 1498–1510.
Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D’Alessio AC et al. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell 2015; 57: 247–260.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567.
Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R et al. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 2011; 108: 14566–14571.
Zhao Z, Chen S, Zhu X, Pan F, Li R, Zhou Y et al. The catalytic activity of TET2 is essential for its myeloid malignancy-suppressive function in hematopoietic stem/progenitor cells. Leukemia 2016; 30: 1784–1788.
Muto H, Sakata-Yanagimoto M, Nagae G, Shiozawa Y, Miyake Y, Yoshida K et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J 2014; 4: e264.
Rasmussen KD, Helin K . Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 2016; 30: 733–750.
Huang H, Jiang X, Li Z, Li Y, Song CX, He C et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA 2013; 110: 11994–11999.
Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol 2015; 16: 653–662.
An J, González-Avalos E, Chawla A, Jeong M, López-Moyado IF, Li W et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 2015; 6: 10071.
Zhao Z, Chen L, Dawlaty MM, Pan F, Weeks O, Zhou Y et al. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep 2015; 13: 1692–1704.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2010; 465: 966.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013; 340: 626–630.
Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 2012; 488: 656–659.
Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 2014; 14: 329–341.
Chen C, Liu Y, Lu C, Cross JR, Morris JP 4th, Shroff AS et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev 2013; 27: 1974–1985.
Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 2010; 467: 338–342.
Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 2012; 47: 633–647.
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2012; 44: 23–31.
Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 2014; 15: 350–364.
Yang L, Rau R, Goodell MA . DNMT3A in haematological malignancies. Nat Rev Cancer 2015; 15: 152–165.
Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet 2016; 48: 1014–1023.
Scourzic L, Couronné L, Pedersen MT, Della Valle V, Diop M, Mylonas E et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia 2016; 30: 1388–1398.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Republic of Korea (NRF-2015R1D1A1A01059801 to MK) and by the Ministry of Science, ICT & Future Planning (MSIP) (no. 2010-0028684 to MK). This work was also supported by the Korea Health Technology Research and Development Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare (HI15C2731 to MK). MK is also supported by Center for Genomic Integrity, Institute for Basic Science (IBS, IBS-R022-D1) and the TJ Park Science Fellowship (2.160019.01), funded by POSCO TJ Park Foundation. JA is supported by the research funds for newly appointed professors of Chonbuk National University in 2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
An, J., Rao, A. & Ko, M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med 49, e323 (2017). https://doi.org/10.1038/emm.2017.5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.5
This article is cited by
-
The interaction between DNA methylation and tumor immune microenvironment: from the laboratory to clinical applications
Clinical Epigenetics (2024)
-
Amino acid metabolism in tumor biology and therapy
Cell Death & Disease (2024)
-
TET3 as a non-invasive screening tool for the detection of fibrosis in patients with chronic liver disease
Scientific Reports (2023)
-
Differential methylation in EGLN1 associates with blood oxygen saturation and plasma protein levels in high-altitude pulmonary edema
Clinical Epigenetics (2022)
-
Targeted erasure of DNA methylation by TET3 drives adipogenic reprogramming and differentiation
Nature Metabolism (2022)