Abstract
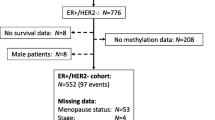
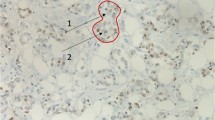
Metastasis tumor antigen 1 (MTA1), a novel candidate metastasis-associated gene, is known to increase the migration and invasion of various tumor cells in vitro. It also plays an important role in tumorigenesis and tumor aggressiveness of breast cancer. Estrogen receptor alpha (ERα) plays an important role in the etiology of breast cancer and has been widely accepted as a prognostic marker for breast cancer and a response predictor for endocrine therapy. The ERα gene methylation has been linked to the lack of ERα expression in breast cancer. The aim of the study is to assess the correlation between the ERα methylation and MTA1 expression in breast cancer and further to investigate whether the repressed ERα methylation can downregulate the expression of MTA1 in vitro. In general, we found ERα methylation had significant correlation with the MTA1 expression (p < 0.05) in female patients of breast cancer (n = 102) by methylation-specific polymerase chain reaction and immunohistochemistry. To gain a deeper insight into the molecular mechanism underlying the relation between MTA1 and ERα methylation, we treated the invasive breast cancer cell lines with the demethylating agent, found the downregulation of MTA1 protein expression, and mRNA with the unmethylation of ERα (p < 0.05). And the invasive ability of breast cancer cells was significantly positively associated with MTA1 expression. These unique findings have greatly extended our current knowledge about the relation between ERα methylation and MTA1 expression. These data strongly support the hypothesis that methylation is involved in the relation between MTA1 and ERα in breast cancer.




Similar content being viewed by others
References
Ghanta KS, Li DQ, Eswaran J, Kumar R. Gene profiling of MTA1 identifies novel gene targets and functions. PLoS One. 2011;6:e17135.
Molli PR, Singh RR, Lee SW, Kumar R. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–80.
Hofer MD, Chang MC, Hirko KA, Rubin MA, Nosé V. Immunohistochemical and clinicopathological correlation of the metastasis-associated gene 1 (MTA1) expression in benign and malignant pancreatic endocrine tumors. Mod Pathol. 2009;22:933–9.
Murakami M, Kaul R, Robertson ES. MTA1 expression is linked to ovarian cancer. Cancer Biol Ther. 2008;7:1468–70.
Tong D, Heinze G, Schremmer M, Schuster E, Czerwenka K, Leodolter S, et al. Expression of the human MTA1 gene in breast cell lines and in breast cancer tissues. Oncol Res. 2007;16:465–70.
Ahmad Z, Khurshid A, Qureshi A, Idress R, Asghar N, Kayani N. Breast carcinoma grading, estimation of tumor size, axillary lymph node status, staging, and Nottingham prognostic index scoring on mastectomy specimens. Indian J Pathol Microbiol. 2009;52:477–81.
Duss S, André S, Nicoulaz AL, Fiche M, Bonnefoi H, Brisken C, et al. An oestrogen-dependent model of breast cancer created by transformation of normal human mammary epithelial cells. Breast Cancer Res. 2007;9:R38.
Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6.
Zhao L, Wang L, Jin F, Ma W, Ren J, Wen X, et al. Silencing of estrogen receptor alpha (ERalpha) gene by promoter hypermethylation is a frequent event in Chinese women with sporadic breast cancer. Breast Cancer Res Treat. 2009;117:253–9.
Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3(1):30–7.
Jiang Q, Zhang H, Zhang P. ShRNA-mediated gene silencing of MTA1 influenced on protein expression of ER alpha, MMP-9, CyclinD1 and invasiveness, proliferation in breast cancer cell lines MDA-MB-231 and MCF-7 in vitro. J Exp Clin Cancer Res. 2011;30:60.
Tavassoli FA. Breast pathology: rationale for adopting the ductal intraepithelial neoplasia (DIN) classification. Nat Clin Pract Oncol. 2005;2:116–7.
Tavassoli FA. Correlation between gene expression profiling-based molecular and morphologic classification of breast cancer. Int J Surg Pathol. 2010;18:167S–9S.
Tavassoli FA. Lobular and ductal intraepithelial neoplasia. Pathologe. 2008;29:107–11.
Jing MX, Mao XY, Li C, Wei J, Liu C, Jin F. Estrogen receptor-alpha promoter methylation in sporadic basal-like breast cancer of Chinese women. Tumour Biol. 2011;32(4):713–9.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6.
Lapidus RG, Nass SJ, Butash KA, Parl FF, Weitzman SA, Graff JG, et al. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res. 1998;58:2515–9.
Kok LF, Lee MY, Tyan YS, Wu TS, Cheng YW, Kung MF, et al. Comparing the scoring mechanisms of p16INK4a immunohistochemistry based on independent nucleic stains and independent cytoplasmic stains in distinguishing between endocervical and endometrial adenocarcinomas in a tissue microarray study. Arch Gynecol Obstet. 2010;281:293–300.
Koo CL, Kok LF, Lee MY, Wu TS, Cheng YW, Hsu JD, et al. Scoring mechanisms of p16INK4a immunohistochemistry based on either independent nucleic stain or mixed cytoplasmic with nucleic expression can significantly signal to distinguish between endocervical and endometrial adenocarcinomas in a tissue microarray study. J Transl Med. 2009;7:25.
Dowling EC, Klabunde C, Patnick J, Ballard-Barbash R. International Cancer Screening Network (ICSN). Breast and cervical cancer screening programme implementation in 16 countries. J Med Screen. 2010;17:139–46.
Gaffan J, Dacre J, Jones A. Educating undergraduate medical students about oncology: a literature review. J Clin Oncol. 2006;24:1932–9.
Brinkman JA, El-Ashry D. ER re-expression and re-sensitization to endocrine therapies in ER-negative breast cancers. J Mammary Gland Biol Neoplasia. 2009;14:67–78.
Manavathi B, Singh K, Kumar R. MTA family of coregulators in nuclear receptor biology and pathology. Nucl Recept Signal. 2007;5:e010.
Kumar R, Wang RA, Mazumdar A, Talukder AH, Mandal M, Yang Z, et al. A naturally occurring MTA1 variant sequesters oestrogen receptor-alpha in the cytoplasm. Nature. 2002;418:654–7.
Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis. 2009;26(3):215–27.
Flanagan J, Kugler S, Waddell N, Johnstone C, Marsh A, Henderson S, et al. DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. Breast Cancer Res. 2010;12(Suppl1):O4.
Yoshida T, Eguchi H, Nakachi K, Tanimoto K, Higashi Y, Suemasu K, et al. Distinct mechanisms of loss of estrogen receptor alpha gene expression in human breast cancer: methylation of the gene and alteration of trans-acting factors. Carcinogenesis. 2000;21:2193–201.
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91.
Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci U S A. 2006;103:6670–5.
Sahab ZJ, Man YG, Semaan SM, Newcomer RG, Byers SW, Sang QX. Alteration in protein expression in estrogen receptor alpha-negative human breast cancer tissues indicates a malignant and metastatic phenotype. Clin Exp Metastasis. 2010;27:493–503.
Acknowledgment
This work was supported by Scientific Research Foundation for the Doctoral Program, Ministry of Science and Technology, Liaoning Province (no. 20121126).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mao, Xy., Chen, H., Wang, H. et al. MTA1 expression correlates significantly with ER-alpha methylation in breast cancer. Tumor Biol. 33, 1565–1572 (2012). https://doi.org/10.1007/s13277-012-0410-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0410-7